We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1072
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
===Catalase Peroxidases=== | ===Catalase Peroxidases=== | ||
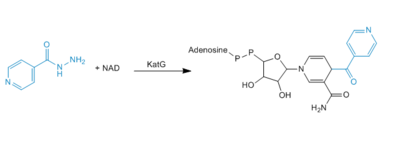
Catalase-peroxidases are enzymes that degrade hydrogen peroxide. Catalase converts two equivalents of hydrogen peroxide into water and oxygen via a two-step reaction cycle in which H<sub>2</sub>0<sub>2</sub> alternately oxidizes and reduces the heme iron at the active site. Within peroxidases, oxidation of heme iron involves a H<sub>2</sub>0<sub>2</sub> molecules, similar to that in the catalase-catalyzed reaction. Reduction of the heme iron, however, involves hydrogen donors such as NADH, not a second H<sub>2</sub>0<sub>2</sub> molcule (3). Catalase-Peroxidases that have been characterized are either homodimers or homotetramers and contain a single heme ''b'' cofactor at the active site. Usually, the primary struture of the subunit can be divided into two halves that have a high level of sequence similarity, most likely due to a gene duplication event. | Catalase-peroxidases are enzymes that degrade hydrogen peroxide. Catalase converts two equivalents of hydrogen peroxide into water and oxygen via a two-step reaction cycle in which H<sub>2</sub>0<sub>2</sub> alternately oxidizes and reduces the heme iron at the active site. Within peroxidases, oxidation of heme iron involves a H<sub>2</sub>0<sub>2</sub> molecules, similar to that in the catalase-catalyzed reaction. Reduction of the heme iron, however, involves hydrogen donors such as NADH, not a second H<sub>2</sub>0<sub>2</sub> molcule (3). Catalase-Peroxidases that have been characterized are either homodimers or homotetramers and contain a single heme ''b'' cofactor at the active site. Usually, the primary struture of the subunit can be divided into two halves that have a high level of sequence similarity, most likely due to a gene duplication event. | ||
| + | |||
==Structure== | ==Structure== | ||
[[Image:DotSTRUCTURE.png|300 px|left|thumb|Overall structure of [http://www.rcsb.org/pdb/explore/explore.do?structureId=1sJ2 1SJ2], showing that the structure is a [http://en.wikipedia.org/wiki/Protein_dimer homodimer].]] | [[Image:DotSTRUCTURE.png|300 px|left|thumb|Overall structure of [http://www.rcsb.org/pdb/explore/explore.do?structureId=1sJ2 1SJ2], showing that the structure is a [http://en.wikipedia.org/wiki/Protein_dimer homodimer].]] | ||
| - | ==Structure== | ||
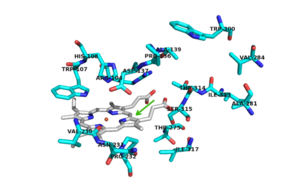
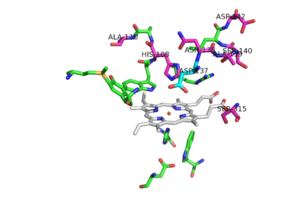
The two [http://en.wikipedia.org/wiki/Protein_domain domains] of each monomer are primarily [http://en.wikipedia.org/wiki/Alpha_helix alpha helical] and have similar foldings. The similar foldings suggests that the monomer results from a [http://en.wikipedia.org/wiki/Gene_duplication gene duplication] event; however, the C-terminal domain does not contain the [http://en.wikipedia.org/wiki/Heme_B heme ''b''] prosthetic group, while the <scene name='69/694238/N_terminus/1'>N terminal</scene> does. The [http://en.wikipedia.org/wiki/Active_site active site] is therefore located within the N-terminal domain. The two monomers interact through an interlocking hook formed by the N-terminal domains that stabilizes the formation of the dimer (1). | The two [http://en.wikipedia.org/wiki/Protein_domain domains] of each monomer are primarily [http://en.wikipedia.org/wiki/Alpha_helix alpha helical] and have similar foldings. The similar foldings suggests that the monomer results from a [http://en.wikipedia.org/wiki/Gene_duplication gene duplication] event; however, the C-terminal domain does not contain the [http://en.wikipedia.org/wiki/Heme_B heme ''b''] prosthetic group, while the <scene name='69/694238/N_terminus/1'>N terminal</scene> does. The [http://en.wikipedia.org/wiki/Active_site active site] is therefore located within the N-terminal domain. The two monomers interact through an interlocking hook formed by the N-terminal domains that stabilizes the formation of the dimer (1). | ||
Revision as of 13:15, 14 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
| |||||||||||