We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1072
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
| - | The two [http://en.wikipedia.org/wiki/Protein_domain domains] of each monomer are primarily [http://en.wikipedia.org/wiki/Alpha_helix alpha helical] and have similar foldings. The similar foldings suggests that the monomer results from a [http://en.wikipedia.org/wiki/Gene_duplication gene duplication] event; however, the C-terminal domain does not contain the [http://en.wikipedia.org/wiki/Heme_B heme ''b''] prosthetic group, while the <scene name='69/694238/N_terminus/1'>N terminal</scene> does. The [http://en.wikipedia.org/wiki/Active_site active site] is therefore located within the N-terminal domain. The two monomers interact through an interlocking hook formed by the N-terminal domains that stabilizes the formation of the dimer | + | The two [http://en.wikipedia.org/wiki/Protein_domain domains] of each monomer are primarily [http://en.wikipedia.org/wiki/Alpha_helix alpha helical] and have similar foldings. The similar foldings suggests that the monomer results from a [http://en.wikipedia.org/wiki/Gene_duplication gene duplication] event; however, the C-terminal domain does not contain the [http://en.wikipedia.org/wiki/Heme_B heme ''b''] prosthetic group, while the <scene name='69/694238/N_terminus/1'>N terminal</scene> does. The [http://en.wikipedia.org/wiki/Active_site active site] is therefore located within the N-terminal domain. The two monomers interact through an interlocking hook formed by the N-terminal domains that stabilizes the formation of the dimer <ref>PMID: 1523184</ref>. |
The N-terminal <scene name='69/694238/N_terminus/2'>hook</scene> is formed through hydrophobic interactions between residues Tyr-28 and Tyr-197 and residues Trp-38 and Trp-204. This interlocking loop region is also found in similar conformations of other catalase peroxidase structures such as: [http://www.proteopedia.org/wiki/index.php/1itk ''hm''CP] and ''bp''CP. | The N-terminal <scene name='69/694238/N_terminus/2'>hook</scene> is formed through hydrophobic interactions between residues Tyr-28 and Tyr-197 and residues Trp-38 and Trp-204. This interlocking loop region is also found in similar conformations of other catalase peroxidase structures such as: [http://www.proteopedia.org/wiki/index.php/1itk ''hm''CP] and ''bp''CP. | ||
| Line 43: | Line 43: | ||
==Mutations== | ==Mutations== | ||
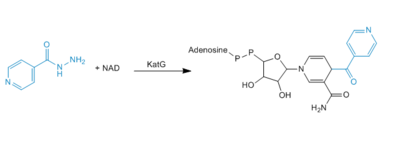
| - | Resistance to INH is a continuing problem in the development of effective therapeutic regimes designed to eliminate infections from ''M. tuberculosis''. Resistance to INH is due to deletions or mutations in this KatG catalase peroxidase enzyme. There are many possible mutations in this peroxidase that can play a role in the resistance of INH. The most commonly occurring mutation occurs at Ser 315. A mutation at this amino acid can result in up to a 200 fold increase in the minimum inhibitory concentration for INH. Ser 315 has been reported to mutate to asparagine, isoleucine, glycine, and most frequently, threonine. A S315T mutant has the ability to reduce the affinity of the enzyme for INH by increasing steric hindrance and reducing access to the substrate binding site | + | Resistance to INH is a continuing problem in the development of effective therapeutic regimes designed to eliminate infections from ''M. tuberculosis''. Resistance to INH is due to deletions or mutations in this KatG catalase peroxidase enzyme. There are many possible mutations in this peroxidase that can play a role in the resistance of INH. The most commonly occurring mutation occurs at Ser 315. A mutation at this amino acid can result in up to a 200 fold increase in the minimum inhibitory concentration for INH. Ser 315 has been reported to mutate to asparagine, isoleucine, glycine, and most frequently, threonine. A S315T mutant has the ability to reduce the affinity of the enzyme for INH by increasing steric hindrance and reducing access to the substrate binding site <ref>PMID: 1523184</ref>. Mutation of Ser315 to a Thr in ''mt''CP results in a loss of the activation to the anti-tuberculosis drug (INH) with no loss of either peroxidase or catalase activity (3). Any of the other mutations at this site, except for glycine, would also increase steric hindrance and decrease the accessibility to the binding site. |
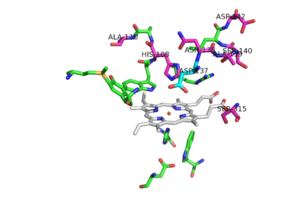
[[Image:Mutation_locations.png|300 px|left|thumb|Pink residues represent the location of possible mutations. Green residues represent the active site. Asp 137 is shown in blue.]] | [[Image:Mutation_locations.png|300 px|left|thumb|Pink residues represent the location of possible mutations. Green residues represent the active site. Asp 137 is shown in blue.]] | ||
| Line 49: | Line 49: | ||
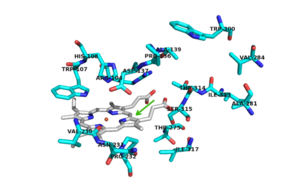
Of the active site residues that are involved in enzyme catalyzed activation of INH, only His 108 has been a site for mutations that can increase resistance to INH. His 108 has been reported to mutate to glutamic acid and [http://en.wikipedia.org/wiki/Glutamine glutamine]. These mutations reduce the affinity for INH but the hydrogen bond donor/acceptor groups of glutamine would still allow INH to bind. However, glutamine wouldn't be able to act as proton shuttle in the way His 108 does in the enzyme-catalyzed activation pathway. | Of the active site residues that are involved in enzyme catalyzed activation of INH, only His 108 has been a site for mutations that can increase resistance to INH. His 108 has been reported to mutate to glutamic acid and [http://en.wikipedia.org/wiki/Glutamine glutamine]. These mutations reduce the affinity for INH but the hydrogen bond donor/acceptor groups of glutamine would still allow INH to bind. However, glutamine wouldn't be able to act as proton shuttle in the way His 108 does in the enzyme-catalyzed activation pathway. | ||
| - | No known mutants have been reported to occur at Asp 137, although a few mutants nearby could cause local conformational changes and thereby altering the orientation of the Asp 137 side chain, making it less effective in binding and activation of INH | + | No known mutants have been reported to occur at Asp 137, although a few mutants nearby could cause local conformational changes and thereby altering the orientation of the Asp 137 side chain, making it less effective in binding and activation of INH <ref>PMID: 1523184</ref>. |
Revision as of 13:37, 14 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
| |||||||||||