Sandbox Reserved 1061
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
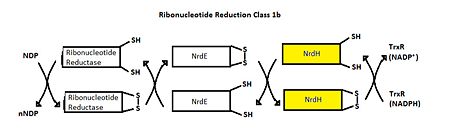
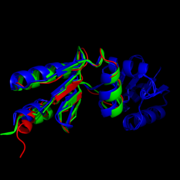
The <scene name='69/694228/Nrdh_structure/1'>MtNrdH structure</scene> determined by x-ray crystallography has 79 residues in a single polypeptide chain. The active site (shown in green) is dominated by a <scene name='69/694228/Nrdh_structure/3'>disulfide bond</scene> between Cys-11 and Cys-14, which serves as the site of reduction by thioredoxin reductase. <ref name="Swastik" /> | The <scene name='69/694228/Nrdh_structure/1'>MtNrdH structure</scene> determined by x-ray crystallography has 79 residues in a single polypeptide chain. The active site (shown in green) is dominated by a <scene name='69/694228/Nrdh_structure/3'>disulfide bond</scene> between Cys-11 and Cys-14, which serves as the site of reduction by thioredoxin reductase. <ref name="Swastik" /> | ||
| - | Many thioredoxin-like proteins have a similar active site region, which includes the <scene name='69/694228/Nrdh_structure/ | + | Many thioredoxin-like proteins have a similar active site region, which includes the |
| + | <scene name='69/694228/Nrdh_structure/6'>thioredoxin fold</scene>, a large turn in the protein structure right before the disulfide bond. The residues directly following the fold, <scene name='69/694228/Nrdh_structure/5'>CVQC</scene>, are the most highly conserved of all areas of the protein across multiple species. | ||
[[Image:Weblogocvqc.png|thumb|center|upright=2.5|Weblogo diagram showing highly conserved CVQC region of NrdH in five separate protein structures from ''Nocardiaseriolae'', ''E. coli'', ''Cornebacterium Ammoniagenes'', and ''Mycobacterium Tuberculosis''.<ref name="weblogo">Crooks GE, Hon G, Chandonia JM, Brenner SE WebLogo: A sequence logo generator, | [[Image:Weblogocvqc.png|thumb|center|upright=2.5|Weblogo diagram showing highly conserved CVQC region of NrdH in five separate protein structures from ''Nocardiaseriolae'', ''E. coli'', ''Cornebacterium Ammoniagenes'', and ''Mycobacterium Tuberculosis''.<ref name="weblogo">Crooks GE, Hon G, Chandonia JM, Brenner SE WebLogo: A sequence logo generator, | ||
Genome Research, 14:1188-1190, (2004)</ref>]] | Genome Research, 14:1188-1190, (2004)</ref>]] | ||
Revision as of 00:28, 22 April 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Structure of Mycobacterium Tuberculosis NrdH
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Swastik, Phulera and Mande, Shekhar C. (2013) The Crystal Structure of Mycobacterium tuberculosis NrdH at 0.87Å Suggests a Possible Mode of Its Activity. Biochemistry 52, 4056-4065.
- ↑ 2.0 2.1 "Tuberculosis." Media Centre. World Health Organization, Web. 16 Mar. 2015. Media Centre. <http://www.who.int/mediacentre/factsheets/fs104/en/>.
- ↑ 3.0 3.1 Crooks GE, Hon G, Chandonia JM, Brenner SE WebLogo: A sequence logo generator, Genome Research, 14:1188-1190, (2004)
- ↑ 4.0 4.1 The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.
- ↑ 5.0 5.1 Kolberg, M., et al. (2004) Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta 1699 (1−2), 1−34.
- ↑ Nelson, David L., and Michael M. Cox. Lehninger Principles of Biochemistry. 5th ed. New York: W.H. Freeman, 2008. 888-889.
- ↑ Makhlynets, O., Boal, A. K., Rhodes, D. V., Kitten, T., Rosenzweig, A. C., & Stubbe, J. (2014). Streptococcus sanguinis Class Ib Ribonucleotide Reductase: HIGH ACTIVITY WITH BOTH IRON AND MANGANESE COFACTORS AND STRUCTURAL INSIGHTS. The Journal of Biological Chemistry, 289(9), 6259–6272. doi:10.1074/jbc.M113.533554.
- ↑ Mowa, M. B., et al. (2009) Function and regulation of class I ribonucleotide reductase-encoding genes in mycobacteria. J. Bacteriol. 191 (3), 985−995