We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1056
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
==Mechanism== | ==Mechanism== | ||
[[Image:Real_Mechanism.png|400 px|right|thumb|Figure 6: Chemical Mechanism of Isocitrate Lyase. Note: The mechanism depicted in this image is the reverse process, but the mechanism is identical.]] | [[Image:Real_Mechanism.png|400 px|right|thumb|Figure 6: Chemical Mechanism of Isocitrate Lyase. Note: The mechanism depicted in this image is the reverse process, but the mechanism is identical.]] | ||
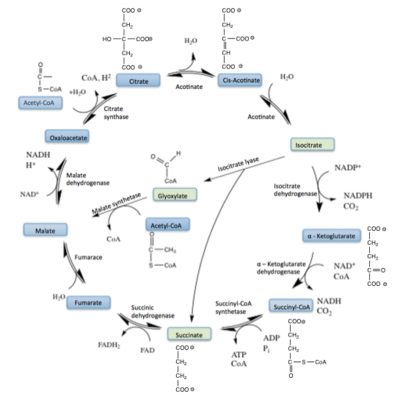
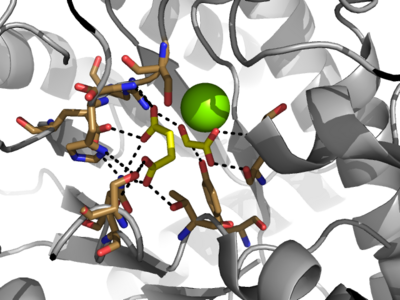
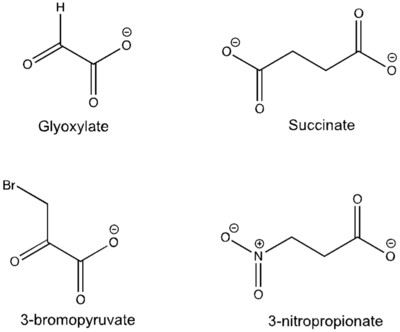
| - | Isocitrate lyase catalyzes a reversible aldol condensation, converting isocitrate to glyoxylate and succinate via the breaking of a C-C bond.<ref name="claisen"/> Within the active site of ICL the HIS193 residue deprotonates the <scene name='69/694223/Arginine/3'>Cys191</scene>residue of the active site in order to increase its basicity.<ref name="ICL">PMID:10932251</ref><ref name="claisen"/> The Cys 191 residue then deprotonates the alpha carbon adjacent to one of the carbonyl groups of succinate, thus forming the enolic intermediate.<ref name="claisen"/> The negatively charged alpha carbon atom of the enolic intermediate acts as a nucleophile that attacks the carbonyl carbon of the aldehyde of glyoxylate. The nucleophilic attack will place a negative charge on the oxygen atom oxygen at the former carbonyl oxygen of the aldehyde, which will be stabilized by positive charges of the Mg ion, ARG228 and HIS180.<ref name="ICL">PMID:10932251</ref> The protonation of this species will yield the final product. It is important to note that this reaction is entirely reversible; the breakdown of isocitrate into glyoxylate and succinate occurs using a similar mechanism. | + | Isocitrate lyase catalyzes a reversible aldol condensation, converting isocitrate to glyoxylate and succinate via the breaking of a C-C bond.<ref name="claisen"/> Within the active site of ICL the HIS193 residue deprotonates the <scene name='69/694223/Arginine/3'>Cys191</scene> residue of the active site in order to increase its basicity.<ref name="ICL">PMID:10932251</ref><ref name="claisen"/> The Cys 191 residue then deprotonates the alpha carbon adjacent to one of the carbonyl groups of succinate, thus forming the enolic intermediate.<ref name="claisen"/> The negatively charged alpha carbon atom of the enolic intermediate acts as a nucleophile that attacks the carbonyl carbon of the aldehyde of glyoxylate. The nucleophilic attack will place a negative charge on the oxygen atom oxygen at the former carbonyl oxygen of the aldehyde, which will be stabilized by positive charges of the Mg ion, ARG228 and HIS180.<ref name="ICL">PMID:10932251</ref> The protonation of this species will yield the final product. It is important to note that this reaction is entirely reversible; the breakdown of isocitrate into glyoxylate and succinate occurs using a similar mechanism. |
Revision as of 03:48, 22 April 2015
Isocitrate Lyase from Mycobacterium Tuberculosis
| |||||||||||
3D Structures of Isocitrate Lyase
Updated on 22-April-2015
- ICL from other bacteria
References
- ↑ Srivastava V, Jain A, Srivastava BS, Srivastava R. Selection of genes of Mycobacterium tuberculosis upregulated during residence in lungs of infected mice. Tuberculosis (Edinb). 2008 May;88(3):171-7. Epub 2007 Dec 3. PMID:18054522 doi:http://dx.doi.org/10.1016/j.tube.2007.10.002
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Sharma V, Sharma S, Hoener zu Bentrup K, McKinney JD, Russell DG, Jacobs WR Jr, Sacchettini JC. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat Struct Biol. 2000 Aug;7(8):663-8. PMID:10932251 doi:10.1038/77964
- ↑ 3.0 3.1 3.2 Beeching JR. High sequence conservation between isocitrate lyase from Escherichia coli and Ricinus communis. Protein Seq Data Anal. 1989 Dec;2(6):463-6. PMID:2696959
- ↑ 4.0 4.1 4.2 4.3 Masamune et al. Bio-Claisen condensation catalyzed by thiolase from Zoogloea ramigera. Active site cysteine residues. "Journal of the American Chemical Society" 111: 1879-1881 (1989). DOI: 10.1021/ja00187a053
- ↑ Connely, M. L. Solvent-accessible surfaces of proteins and nucleic acids "Science" 221:709-713 (1983). DOI: 10.1126/science.6879170