This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1087
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== '''AcrB Transporter''' == | == '''AcrB Transporter''' == | ||
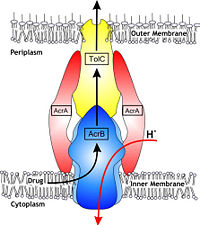
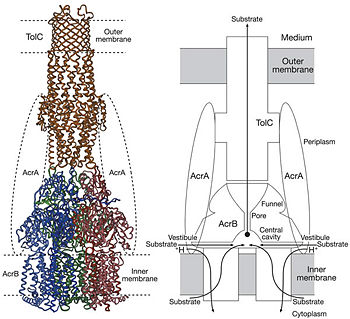
| - | = | + | [[Image:efflux pump.jpg|thumb|left|200px|Figure 1. Hypothetical structure of tripartite efflux pumps in Gram-negative bacteria, e.g. the AcrA/AcrB/TolC efflux pump from ''E. coli''<ref name= "Nakashima"/>]] |
| - | + | In Gram-negative bacteria, such as Escherichia coli, one of the main reasons behind bacterial multidrug resistance is resistance nodulation cell division (RND) transporters which pump a wide range of dyes, bile salts, detergents and structurally unrelated antibiotics out of the cell by proton motive force driven efflux. The natural function of the efflux system is currently under debate. AcrB is the major RND transporter in E. coli. Three proteins form the tripartite multidrug efflux system that pumps the drugs out from the cell. The AcrB transporter, the inner membrane component of the system, cooperates with two other proteins: membrane fusion protein AcrA and an outer membrane channel TolC. The TolC component is connected to outer membrane and AcrB is connected to the inner membrane. The AcrA connects TolC and AcrB together (Fig 1). The structure of the complex indicates that the drugs are transported out of the cell in a three-step procedure.<ref name= "Nakashima">PMID: 12607261</ref><ref name= "Klaas">PMID: 19166984</ref><ref name= "Murasaki">PMID: 18644451</ref> | |
| - | |||
| - | |||
| - | In Gram-negative bacteria, such as Escherichia coli, one of the main reasons behind bacterial multidrug resistance is resistance nodulation cell division (RND) transporters which pump a wide range of dyes, bile salts, detergents and structurally unrelated antibiotics out of the cell by proton motive force driven efflux. The natural function of the efflux system is currently under debate. AcrB is the major RND transporter in E. coli. Three proteins form the tripartite multidrug efflux system that pumps the drugs out from the cell. The AcrB transporter, the inner membrane component of the system, cooperates with two other proteins: membrane fusion protein AcrA and an outer membrane channel TolC. The TolC component is connected to outer membrane and AcrB is connected to the inner membrane. The AcrA connects TolC and AcrB together (Fig 1). The structure of the complex indicates that the drugs are transported out of the cell in a three-step procedure.<ref name= "Nakashima">PMID: 12607261</ref><ref name= "Klaas">PMID: 19166984</ref><ref name= "Murasaki">PMID: 18644451</ref> | ||
| Line 30: | Line 27: | ||
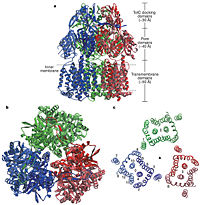
[[Image:Nature01050-f2.2.jpg|thumb|left|200px|Figure 2. Structure of AcrB. a) Side view of the ribbon representation. The three protomers are | [[Image:Nature01050-f2.2.jpg|thumb|left|200px|Figure 2. Structure of AcrB. a) Side view of the ribbon representation. The three protomers are | ||
| - | individually coloured (blue, green and red), b) Top view of a ribbon representation, c) Structure within a slab of the transmembrane domain parallel to the membrane plane near the periplasmic surface.]] | + | individually coloured (blue, green and red), b) Top view of a ribbon representation, c) Structure within a slab of the transmembrane domain parallel to the membrane plane near the periplasmic surface.<ref name= "Nakashima"/>]] |
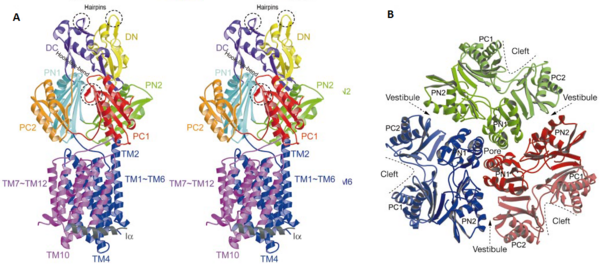
| - | [[Image:Figure5.png|thumb|right|600px|Figure 3. The structure of a single protomer. A) Topology diagram of the protomer, B) A stereo view of ribbon representation of a protomer viewed from outside the headpiece]] | + | [[Image:Figure5.png|thumb|right|600px|Figure 3. The structure of a single protomer. A) Topology diagram of the protomer, B) A stereo view of ribbon representation of a protomer viewed from outside the headpiece<ref name= "Nakashima"/>]] |
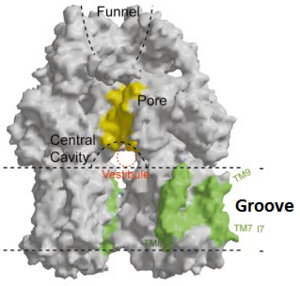
| - | [[Image:Fig groove.png|thumb|left|300px|Figure 4. The groove in the transmembrane domain highlighted in green]] | + | [[Image:Fig groove.png|thumb|left|300px|Figure 4. The groove in the transmembrane domain highlighted in green<ref name= "Nakashima"/>]] |
In the AcrB, there is three domains: TolC docking domain, pore domain and the transmembrane domain. The AcrB consists of 1 049 amino acids and the three monomers of AcrB are organized as a trimer (Fig. 2). The first structure of the trimer was derived in symmetric manner <scene name='70/700000/Acrb_trimer/1'>trimer</scene> (Fig. 2) at 3.5 Å resolution. Later, a new 2.9 Å asymmetric conformation of trimeric AcrB was obtained and three structurally different monomers were spotted<ref>PMID: 16946072</ref>. The appearance of the trimer is of a jellyfish. The N- and C-terminal half’s show similar structural architecture and indicates at early gene duplication events. The trimer, formed by AcrB monomers, appears to be stabilized by the intermonomer connecting loops.<ref name= "Nakashima"/> | In the AcrB, there is three domains: TolC docking domain, pore domain and the transmembrane domain. The AcrB consists of 1 049 amino acids and the three monomers of AcrB are organized as a trimer (Fig. 2). The first structure of the trimer was derived in symmetric manner <scene name='70/700000/Acrb_trimer/1'>trimer</scene> (Fig. 2) at 3.5 Å resolution. Later, a new 2.9 Å asymmetric conformation of trimeric AcrB was obtained and three structurally different monomers were spotted<ref>PMID: 16946072</ref>. The appearance of the trimer is of a jellyfish. The N- and C-terminal half’s show similar structural architecture and indicates at early gene duplication events. The trimer, formed by AcrB monomers, appears to be stabilized by the intermonomer connecting loops.<ref name= "Nakashima"/> | ||
| Line 55: | Line 52: | ||
== Function == | == Function == | ||
| - | [[Image:Nature01050-f5.2.jpg|thumb|right|350px|Figure 4. Proposed model of the AcrB/AcrA/TolC complex and the schematic mechanism of multidrug export mediated by AcrAB/TolC system]] | + | [[Image:Nature01050-f5.2.jpg|thumb|right|350px|Figure 4. Proposed model of the AcrB/AcrA/TolC complex and the schematic mechanism of multidrug export mediated by AcrAB/TolC system<ref name= "Nakashima"/>]] |
Possible mechanism of transport function has been postulated. ArcB can cooperate with TolC in the TolC docking domain forming a direct pathway from the cytoplasm to the extracellular milieu. Based on the information of structure, there might be two pathways for the substrate translocation to the cavity. One is the groove located between TM8 and TM7 in the transmembrane domain of each monoter (Fig 1C). The other is the vestibules which are between PN2 and PC2 of the pore domains. The substrates located in the inner leaflet of the membrane and cytoplasm would get access to the cavity through the transmembrane groove, while the substrates that are on the outer space or in the outer leaflet of the membrane are more likely to be transported to the cavity through the vestibules.<ref name= "Nakashima"/> | Possible mechanism of transport function has been postulated. ArcB can cooperate with TolC in the TolC docking domain forming a direct pathway from the cytoplasm to the extracellular milieu. Based on the information of structure, there might be two pathways for the substrate translocation to the cavity. One is the groove located between TM8 and TM7 in the transmembrane domain of each monoter (Fig 1C). The other is the vestibules which are between PN2 and PC2 of the pore domains. The substrates located in the inner leaflet of the membrane and cytoplasm would get access to the cavity through the transmembrane groove, while the substrates that are on the outer space or in the outer leaflet of the membrane are more likely to be transported to the cavity through the vestibules.<ref name= "Nakashima"/> | ||
Revision as of 13:23, 23 April 2015
| |||||||||||