We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1063
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
The active site of FadD13 is composed of an <scene name='69/694230/Fadd13_subunits/12'>ATP/AMP binding region</scene>. This region is comprised of residues 164-TSGTTGHPKG173-173 shown in red (show atoms by color?) which binds to the phosphate group, and residues 298-VQGYALTE-305 shown in blue which binds to the adenine group. Research has also shown that <scene name='69/694230/Fadd13_subunits/16'>Ser 404</scene> plays a major role in binding of CoA. Ser 404 was shown to have a 4-fold enhancement for the Km value of CoA.<ref name="Khare 2009">DOI: 10.1371/journal.pone.0008387</ref> | The active site of FadD13 is composed of an <scene name='69/694230/Fadd13_subunits/12'>ATP/AMP binding region</scene>. This region is comprised of residues 164-TSGTTGHPKG173-173 shown in red (show atoms by color?) which binds to the phosphate group, and residues 298-VQGYALTE-305 shown in blue which binds to the adenine group. Research has also shown that <scene name='69/694230/Fadd13_subunits/16'>Ser 404</scene> plays a major role in binding of CoA. Ser 404 was shown to have a 4-fold enhancement for the Km value of CoA.<ref name="Khare 2009">DOI: 10.1371/journal.pone.0008387</ref> | ||
===Hydrophobic Tunnel=== | ===Hydrophobic Tunnel=== | ||
| - | FadD13 has a distinct hydrophobic tunnel that starts at the active site and is capped by a positively charged surface patch.The <scene name='69/694230/Fadd13_subunits/13'>hydrophobic tunnel</scene> is found inside the N-terminal domain. It is composed of six beta sheets (beta 9-14) shown in green and two alpha helices (alpha 8-9) shown in red.The hydrophobic tunnel allows large lipids/fatty acids, up to 26 carbons, to bind. | + | FadD13 has a distinct hydrophobic tunnel that starts at the active site and is capped by a positively charged surface patch.The <scene name='69/694230/Fadd13_subunits/13'>hydrophobic tunnel</scene> is found inside the N-terminal domain. It is composed of six beta sheets (beta 9-14) shown in green and two alpha helices (alpha 8-9) shown in red.The hydrophobic tunnel allows large lipids/fatty acids, up to 26 carbons, to bind to the enzyme to be activated. |
===Surface Patch=== | ===Surface Patch=== | ||
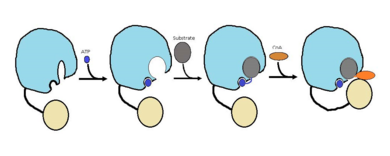
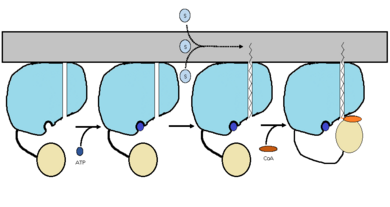
FadD13 has an arginine and aromatic rich surface patch that allows it to be a [http://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral-membrane protein]<ref name="Khare 2009"/>.The hydrophobic tunnel is capped by the arginine and aromatic rich <scene name='69/694230/Fadd13_subunits/14'>lid loop</scene> shown in yellow that is involved in the peripheral binding of the enzyme to the membrane. Based on the structural information present and biochemical information, it is likely that the lid loop opens up upon contact with the membrane. This would allow for the substrate to bind and have the lipid tail to reside in the membrane during catalysis (figure 2)<ref name="Anderson 2012"/>. Six key arginine residues, <scene name='69/694230/Fadd13_subunits/15'>Arg 9, 17, 195, 197, 199, and 244</scene> create a positively charged surface that is likely involved in initially recruiting FadD13 to the membrane. When these residues were replaced with hyrdrophobic alanine residues, membrane binding increased. This points to the important role of hydrophobic interactions in keeping the protein bound at the membrane<ref name="Anderson 2012">PMID: 22560731</ref>. | FadD13 has an arginine and aromatic rich surface patch that allows it to be a [http://en.wikipedia.org/wiki/Peripheral_membrane_protein peripheral-membrane protein]<ref name="Khare 2009"/>.The hydrophobic tunnel is capped by the arginine and aromatic rich <scene name='69/694230/Fadd13_subunits/14'>lid loop</scene> shown in yellow that is involved in the peripheral binding of the enzyme to the membrane. Based on the structural information present and biochemical information, it is likely that the lid loop opens up upon contact with the membrane. This would allow for the substrate to bind and have the lipid tail to reside in the membrane during catalysis (figure 2)<ref name="Anderson 2012"/>. Six key arginine residues, <scene name='69/694230/Fadd13_subunits/15'>Arg 9, 17, 195, 197, 199, and 244</scene> create a positively charged surface that is likely involved in initially recruiting FadD13 to the membrane. When these residues were replaced with hyrdrophobic alanine residues, membrane binding increased. This points to the important role of hydrophobic interactions in keeping the protein bound at the membrane<ref name="Anderson 2012">PMID: 22560731</ref>. | ||
Revision as of 00:52, 24 April 2015
FadD13
| |||||||||||
References
- ↑ Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J Lipid Res. 2007 Dec;48(12):2736-50. Epub 2007 Aug 30. PMID:17762044 doi:http://dx.doi.org/M700378-JLR200
- ↑ Kochan G, Pilka ES, von Delft F, Oppermann U, Yue WW. Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A. J Mol Biol. 2009 May 22;388(5):997-1008. Epub 2009 Apr 1. PMID:19345228 doi:10.1016/j.jmb.2009.03.064
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Andersson CS, Lundgren CA, Magnusdottir A, Ge C, Wieslander A, Molina DM, Hogbom M. The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme. Structure. 2012 May 2. PMID:22560731 doi:10.1016/j.str.2012.03.012
- ↑ 4.0 4.1 4.2 Khare G, Gupta V, Gupta RK, Gupta R, Bhat R, Tyagi AK. Dissecting the role of critical residues and substrate preference of a Fatty Acyl-CoA Synthetase (FadD13) of Mycobacterium tuberculosis. PLoS One. 2009 Dec 21;4(12):e8387. doi: 10.1371/journal.pone.0008387. PMID:20027301 doi:10.1371/journal.pone.0008387