Sandbox Reserved 1059
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
==Structure== | ==Structure== | ||
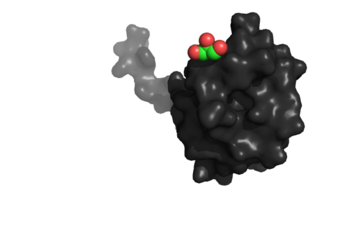
| - | [[Image:Image_2_(2).png|350px|left|thumb|Binding site specific for aromatic amino acids. The hole that is located in the center of this image of NrdH shows the binding site.]] | + | [[Image:Image_2_(2).png|350px|left|thumb|'''Figure 1:'''Binding site specific for aromatic amino acids. The hole that is located in the center of this image of NrdH shows the binding site.]] |
The <scene name='69/694226/Tertiary_structure/4'>tertiary structure</scene> of NrdH has a thioredoxin fold with 79 residues with a glutaredoxin-like sequence. Unlike glutaredoxins, NrdH of ''Mycobacterium tuberculosis'' can accept electrons from thioredoxin reductase<ref name="Phulera" />. It contains two bound ligands, three alpha helices, and four beta sheets. The two ligands are colored by element, red representing oxygen and gray representing carbon. The binding site of NrdH is specific for aromatic amino acids. The image on the left shows the specific binding site. The specificity for aromatic amino acids is vital because aromatic-aromatic interactions have shown to have great importance for protein folding, ligand binding, and protein stability <ref name="Lanzarotti">DOI: 10.1021/ci200062e</ref>. | The <scene name='69/694226/Tertiary_structure/4'>tertiary structure</scene> of NrdH has a thioredoxin fold with 79 residues with a glutaredoxin-like sequence. Unlike glutaredoxins, NrdH of ''Mycobacterium tuberculosis'' can accept electrons from thioredoxin reductase<ref name="Phulera" />. It contains two bound ligands, three alpha helices, and four beta sheets. The two ligands are colored by element, red representing oxygen and gray representing carbon. The binding site of NrdH is specific for aromatic amino acids. The image on the left shows the specific binding site. The specificity for aromatic amino acids is vital because aromatic-aromatic interactions have shown to have great importance for protein folding, ligand binding, and protein stability <ref name="Lanzarotti">DOI: 10.1021/ci200062e</ref>. | ||
| Line 17: | Line 17: | ||
Members of the NrdH family are typically characterized by CVQC and WSGFRP <scene name='69/694226/Conserved_motifs/2'>conserved sequence motifs</scene>. | Members of the NrdH family are typically characterized by CVQC and WSGFRP <scene name='69/694226/Conserved_motifs/2'>conserved sequence motifs</scene>. | ||
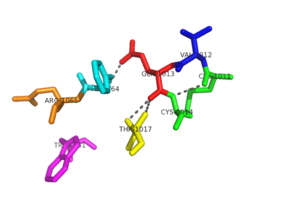
| - | [[Image:Conserved Motifs.png|300px|left|thumb|The two conserved sequence motifs: CVQC and WSGFRP have a network of hydrogen bonding that stabilizes the redox active site in NrdH. This hydrogen bonding network allows for specific interactions with substrates.]] | + | [[Image:Conserved Motifs.png|300px|left|thumb|'''Figure 2:'''The two conserved sequence motifs: CVQC and WSGFRP have a network of hydrogen bonding that stabilizes the redox active site in NrdH. This hydrogen bonding network allows for specific interactions with substrates.]] |
The <scene name='69/694226/Cvqc_motif/3'>CVQC motif</scene>, is an active site and it is located at the N terminus of the first alpha helix<ref name="Laer" />. It is one of the best characterized redox motifs within the thioredoxin-like proteins. The N-terminal cysteine acts as a nucleophile and the C-terminal cysteine acts as the resolving cysteine. Valine is known to be exposed to the solvent. The hydrogen bonding network is important for stability to the redox active site <ref name="Phulera" />. | The <scene name='69/694226/Cvqc_motif/3'>CVQC motif</scene>, is an active site and it is located at the N terminus of the first alpha helix<ref name="Laer" />. It is one of the best characterized redox motifs within the thioredoxin-like proteins. The N-terminal cysteine acts as a nucleophile and the C-terminal cysteine acts as the resolving cysteine. Valine is known to be exposed to the solvent. The hydrogen bonding network is important for stability to the redox active site <ref name="Phulera" />. | ||
| Line 45: | Line 45: | ||
==Relevance== | ==Relevance== | ||
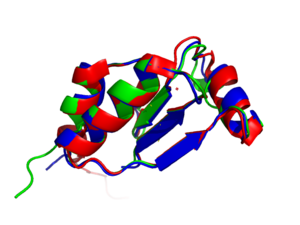
| - | [[Image:Image_7_(1).png|300px|right|thumb|Sequence alignment of NrdH from ''Mycobacterium tuberculosis'', ''Corynebacterium glutamicum'', and ''Echerichia coli'']] | + | [[Image:Image_7_(1).png|300px|right|thumb|'''Figure 3:'''Sequence alignment of NrdH from ''Mycobacterium tuberculosis'', ''Corynebacterium glutamicum'', and ''Echerichia coli''. The thioredoxin fold found in NrdH of ''M. tuberculosis''and it is similar to NrdH of other organisms.]] |
| - | Genes that encode for NrdE and NrdF are essential for growth, and RR might be an attractive biochemical pathway for antimycobacterial drug discovery. Organisms that depend solely on class | + | NrdH of ''M. tuberculosis'' has a thioredoxin fold, which was predicted due to the fact that NrdH of other organisms have a similar thioredoxin fold. This was shown by superimposing the structure of NrdH of multiple organisms. These superimpositions are important because it allows for analyzation of the similarities and differences of NrdH of ''M. tuberculosis'' with glutaredoxin and thioredoxin. There are slight changes in the series of sequence which in turn leads to a change in the tertiary structure. In the process of modeling NrdH with the glutaredoxin-1b RNR C-terminal peptide complex, it was apparent that the peptide bonds were slightly different within the class 1b RNRs. This also provided more evidence for the specificity of NrdH to NrdE. Genes that encode for NrdE and NrdF are essential for growth, and RR might be an attractive biochemical pathway for antimycobacterial drug discovery. Organisms that depend solely on class 1b RNR could potentially contain the essential genes and serve as potential drug targets for treating tuberculosis <ref name="Phulera" />. |
==References== | ==References== | ||
<references/> | <references/> | ||
</StructureSection> | </StructureSection> | ||
Revision as of 03:13, 27 April 2015
NrdH of Mycobacterium tuberculosis
| |||||||||||