Sandbox Reserved 1068
From Proteopedia
| Line 24: | Line 24: | ||
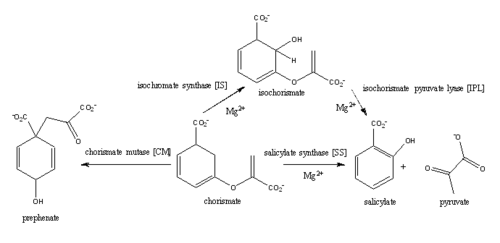
MbtI structure has a mobile element (residues 323 to 327) that can adopt a <scene name='69/694235/Irp9_closed_state/2'>closed</scene> or <scene name='69/694235/2g5f_with_open_loop/1'>open conformation</scene> depending on whether or not ligands are bound to the active site(Harrison 2006). The closed conformation partially obstructs the active site. <ref name= "5a"/>. | MbtI structure has a mobile element (residues 323 to 327) that can adopt a <scene name='69/694235/Irp9_closed_state/2'>closed</scene> or <scene name='69/694235/2g5f_with_open_loop/1'>open conformation</scene> depending on whether or not ligands are bound to the active site(Harrison 2006). The closed conformation partially obstructs the active site. <ref name= "5a"/>. | ||
| - | Inhibition studies have also shown a switch in binding mode at the MbtI active site for inhibitors carrying a substituted enolpyruvyl group compared to the chorismate substrate(Chi 2006, Turvey 2012, Turvey 2010). Crystal structures and fluorescent-based thermal shift assays show that substituents larger than a methyl group are accommodated in the active site of MbtI through localized flexibility in the peptide backbone(Chi 2006). Positioning of | + | Inhibition studies have also shown a switch in binding mode at the MbtI active site for inhibitors carrying a substituted enolpyruvyl group compared to the chorismate substrate(Chi 2006, Turvey 2012, Turvey 2010). Crystal structures and fluorescent-based thermal shift assays show that substituents larger than a methyl group are accommodated in the active site of MbtI through localized flexibility in the peptide backbone(Chi 2006). Positioning of the <scene name='69/694235/3st6_structure_bindingpocket/3'>active site residues</scene> of MbtI in [[3ST6]] with the inhibitor AMT bound is highly similar to the positioning of the <scene name='69/694235/3log_bindingpocket/2'>active site residues</scene> in closed form of MbtI [[3log]] with succinic acid bound <ref name= "5a"/>. |
==Molecular Mechanism== | ==Molecular Mechanism== | ||
Revision as of 17:49, 26 April 2015
Contents |
Mycobacterium tuberculosis salicylate synthase (Mbt1)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Chi G, Manos-Turvey A, O'Connor PD, Johnston JM, Evans GL, Baker EN, Payne RJ, Lott JS, Bulloch EM. Implications of Binding Mode and Active Site Flexibility for Inhibitor Potency against the Salicylate Synthase from Mycobacterium tuberculosis. Biochemistry. 2012 Jun 7. PMID:22607697 doi:10.1021/bi3002067
- ↑ 2.0 2.1 doi: https://dx.doi.org/10.1002/cmdc/201000137
- ↑ 3.0 3.1 Manos-Turvey A, Cergol KM, Salam NK, Bulloch EM, Chi G, Pang A, Britton WJ, West NP, Baker EN, Lott JS, Payne RJ. Synthesis and evaluation of M. tuberculosis salicylate synthase (MbtI) inhibitors designed to probe plasticity in the active site. Org Biomol Chem. 2012 Dec 14;10(46):9223-36. doi: 10.1039/c2ob26736e. Epub 2012, Oct 29. PMID:23108268 doi:http://dx.doi.org/10.1039/c2ob26736e
- ↑ Voss, James J., Kerry Rutter, Benjamin G. Schroedor, Hua Su, and YaQi Zhu. "The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages." Proceedings of the National Academy of Sciences 97.3 (2000): 1252-57. Web. 14 Mar. 2015.
- ↑ Lamb AL. Pericyclic reactions catalyzed by chorismate-utilizing enzymes. Biochemistry. 2011 Sep 6;50(35):7476-83. doi: 10.1021/bi2009739. Epub 2011 Aug, 12. PMID:21823653 doi:http://dx.doi.org/10.1021/bi2009739
- ↑ He Z, Stigers Lavoie KD, Bartlett PA, Toney MD. Conservation of mechanism in three chorismate-utilizing enzymes. J Am Chem Soc. 2004 Mar 3;126(8):2378-85. PMID:14982443 doi:http://dx.doi.org/10.1021/ja0389927
- ↑ Ferrer S, Marti S, Moliner V, Tunon I, Bertran J. Understanding the different activities of highly promiscuous MbtI by computational methods. Phys Chem Chem Phys. 2012 Mar 14;14(10):3482-9. doi: 10.1039/c2cp23149b. Epub, 2012 Feb 3. PMID:22307014 doi:http://dx.doi.org/10.1039/c2cp23149b
- ↑ 8.0 8.1 8.2 8.3 Nicoloff H, Arsene-Ploetze F, Malandain C, Kleerebezem M, Bringel F. Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J Bacteriol. 2004 Sep;186(18):6059-69. PMID:15342575 doi:http://dx.doi.org/10.1128/JB.186.18.6059-6069.2004
- ↑ Ferrer S, Marti S, Moliner V, Tunon I, Bertran J. Understanding the different activities of highly promiscuous MbtI by computational methods. Phys Chem Chem Phys. 2012 Mar 14;14(10):3482-9. doi: 10.1039/c2cp23149b. Epub, 2012 Feb 3. PMID:22307014 doi:http://dx.doi.org/10.1039/c2cp23149b
- ↑ Ferrer S, Marti S, Moliner V, Tunon I, Bertran J. Understanding the different activities of highly promiscuous MbtI by computational methods. Phys Chem Chem Phys. 2012 Mar 14;14(10):3482-9. doi: 10.1039/c2cp23149b. Epub, 2012 Feb 3. PMID:22307014 doi:http://dx.doi.org/10.1039/c2cp23149b
- ↑ He Z, Stigers Lavoie KD, Bartlett PA, Toney MD. Conservation of mechanism in three chorismate-utilizing enzymes. J Am Chem Soc. 2004 Mar 3;126(8):2378-85. PMID:14982443 doi:http://dx.doi.org/10.1021/ja0389927
- ↑ Ferrer S, Marti S, Moliner V, Tunon I, Bertran J. Understanding the different activities of highly promiscuous MbtI by computational methods. Phys Chem Chem Phys. 2012 Mar 14;14(10):3482-9. doi: 10.1039/c2cp23149b. Epub, 2012 Feb 3. PMID:22307014 doi:http://dx.doi.org/10.1039/c2cp23149b
- ↑ Ferrer S, Marti S, Moliner V, Tunon I, Bertran J. Understanding the different activities of highly promiscuous MbtI by computational methods. Phys Chem Chem Phys. 2012 Mar 14;14(10):3482-9. doi: 10.1039/c2cp23149b. Epub, 2012 Feb 3. PMID:22307014 doi:http://dx.doi.org/10.1039/c2cp23149b
- ↑ Tuberculosis (TB). Ed. Sam Posner. Centers for Disease Control and Prevention, n.d. Web. 9 Apr. 2015.
- ↑ De Voss, James J., Kerry Rutter, Benjamin G. Schroeder, Hua Su, and YaQi Zhu. The salicylate-derived mycobacterium siderophore of Mycobacterium tuberculosis are essential for growth in macrophages. "Proceedings of the National Science Academy" 97.3 (2000): 1252-57. Web. 5 Apr. 2015.
Student contributors
Stephanie Raynor and Robin Gagnon
Related pdb files and proteopedia pages
3D structures of isochorismate pyruvate lyase
3log – MtIPL/isochorismate synthase - Mycobacterium tuberculosis
3rv6, 3rv7, 3rv8, 3rv9, 3st6, 3veh - MtIPL/isochorismate synthase + inhibitor
2h9c – PaIPL residues 1-99 – Pseudomonas aeruginosa
2h9d - PaIPL + pyruvate
3LOG
3D structure of isochorismate synthase
2eua, 3bzm, 3bzn - MenF from E. coli
3os6 - DhbC from Bacillus anthracis
3gse - MenF from Yersinia pestis
3hwo - EntC
3D structure of salicylate synthase
3veh - MbtI with inhibitor methylAMT
3st6 - MbtI with isochorismate analogue inhibitor
3rv6 (Phenyl R-group), 3rv7 (Isopropyl R-group), 3rv8 (Cyclopropyl R-group), 3rv9 (Ethyl R-group) - MbtI with inhibitor
2fn0, 2fn1 (with products salicylate and pyruvate) - Irp9 from Yersinia enterocolitica
2i6y - MbtI