Sandbox Reserved 1068
From Proteopedia
| Line 15: | Line 15: | ||
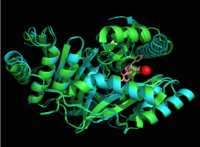
The crystal asymmetric unit was found to contain <scene name='69/694235/3log/11'> four MbtI molecules</scene>, however crystal packing and size exclusion chromatography data suggest a monomeric enzyme <ref name= "3a">PMID 16923875</ref>. There are no significant structural changes between the four monomers excepts from the localized differences in the active site <ref name= "3a"/>. The overall molecular structure consist of a polypeptide of 450 residues that forms <scene name='69/694235/Alpha_helics/2'>one large single domain</scene> with a similar fold to other chromate-utilizing enzymes <ref name="3a"/>. The core of the protein is formed by <scene name='69/694234/Beta_sheets/1'>21 beta sheets </scene>folded into a twisted beta-sandwich. The protein's core is then surrounded by <scene name='69/694235/Beta_sheets/4'>10 alpha helices</scene><ref name="3a"/>. The active site was identified by comparison to the product bound forms of [[Irp9]] and [[TrpE]] and is situated in a cleft that is about 12Å in length, 10Å deep, and 7Å wide <ref name="3a"/>. One side of the groove is formed by β21, C-terminal helix, and α11 while the other side of the groove is formed by β16-17 loop, helix α7, and β15-α6 loop (Figure 2)<ref name="3a"/>. The β19-20 and β12-13 loops make up the bottom of the active side cleft (Figure 2) <ref name="3a">PMID:16923875</ref>. | The crystal asymmetric unit was found to contain <scene name='69/694235/3log/11'> four MbtI molecules</scene>, however crystal packing and size exclusion chromatography data suggest a monomeric enzyme <ref name= "3a">PMID 16923875</ref>. There are no significant structural changes between the four monomers excepts from the localized differences in the active site <ref name= "3a"/>. The overall molecular structure consist of a polypeptide of 450 residues that forms <scene name='69/694235/Alpha_helics/2'>one large single domain</scene> with a similar fold to other chromate-utilizing enzymes <ref name="3a"/>. The core of the protein is formed by <scene name='69/694234/Beta_sheets/1'>21 beta sheets </scene>folded into a twisted beta-sandwich. The protein's core is then surrounded by <scene name='69/694235/Beta_sheets/4'>10 alpha helices</scene><ref name="3a"/>. The active site was identified by comparison to the product bound forms of [[Irp9]] and [[TrpE]] and is situated in a cleft that is about 12Å in length, 10Å deep, and 7Å wide <ref name="3a"/>. One side of the groove is formed by β21, C-terminal helix, and α11 while the other side of the groove is formed by β16-17 loop, helix α7, and β15-α6 loop (Figure 2)<ref name="3a"/>. The β19-20 and β12-13 loops make up the bottom of the active side cleft (Figure 2) <ref name="3a">PMID:16923875</ref>. | ||
| + | For further structural and sequence information see [http://www.uniprot.org/uniprot/P9WFX1]. | ||
== Structural highlights == | == Structural highlights == | ||
| Line 57: | Line 58: | ||
[http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis] is the causative agent of [http://www.cdc.gov/tb/ Tuberculosis] (TB), an infectious disease that affects one-third of the worlds population. Two TB-related conditions exist: latent TB infection and active TB disease. Currently, there are four regimens that are approved for the treatment of latent TB infection through the use of the antibiotics isoniazid, rifampin, and rifapentine.TB disease can also be treated through various antibiotic regimens. There are 10 drugs currently approved by the FDA for treating TB disease. The first-line anti-TB agents are the antibiotics isoniazid, rifampin, ethambutol, and pyrazinamide <ref>Tuberculosis (TB). Ed. Sam Posner. Centers for Disease Control and Prevention, n.d. Web. 9 Apr. 2015.</ref>. Although various treatments for TB infection and TB disease exist, the emergence of [http://www.cdc.gov/tb/publications/factsheets/drtb/mdrtb.htm multi-drug] and [http://www.cdc.gov/tb/topic/drtb/xdrtb.htm extensively-drug] resistant strains of ''M. tuberculosis'' has increased the need for anti-tubercular agents with novel modes of action. | [http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis] is the causative agent of [http://www.cdc.gov/tb/ Tuberculosis] (TB), an infectious disease that affects one-third of the worlds population. Two TB-related conditions exist: latent TB infection and active TB disease. Currently, there are four regimens that are approved for the treatment of latent TB infection through the use of the antibiotics isoniazid, rifampin, and rifapentine.TB disease can also be treated through various antibiotic regimens. There are 10 drugs currently approved by the FDA for treating TB disease. The first-line anti-TB agents are the antibiotics isoniazid, rifampin, ethambutol, and pyrazinamide <ref>Tuberculosis (TB). Ed. Sam Posner. Centers for Disease Control and Prevention, n.d. Web. 9 Apr. 2015.</ref>. Although various treatments for TB infection and TB disease exist, the emergence of [http://www.cdc.gov/tb/publications/factsheets/drtb/mdrtb.htm multi-drug] and [http://www.cdc.gov/tb/topic/drtb/xdrtb.htm extensively-drug] resistant strains of ''M. tuberculosis'' has increased the need for anti-tubercular agents with novel modes of action. | ||
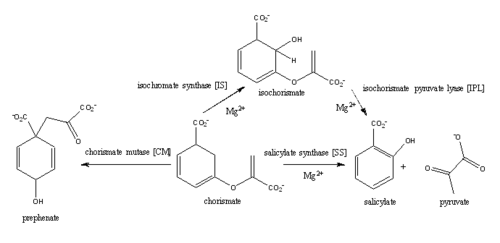
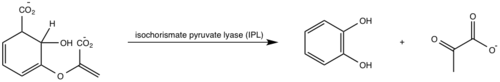
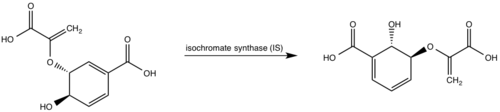
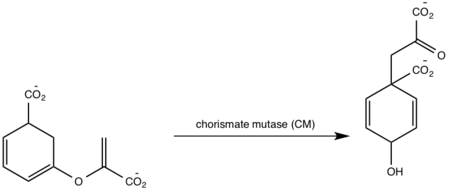
| - | [http://en.wikipedia.org/wiki/Iron#Biological_role Iron] is essential for mycobacterial growth and pathogenesis, therefore the pathways for iron acquisition are potential targets for antibacterial therapies.''M. tuberculosis'' obtains iron through two different pathways: chelating iron from the host through the siderophore mycobactin and the degradation of heme released from damaged red blood cells | + | [http://en.wikipedia.org/wiki/Iron#Biological_role Iron] is essential for mycobacterial growth and pathogenesis, therefore the pathways for iron acquisition are potential targets for antibacterial therapies.''M. tuberculosis'' obtains iron through two different pathways: chelating iron from the host through the siderophore mycobactin and the degradation of heme released from damaged red blood cells<ref name="1a"/>. MbtI catalyses the first committed step in the biosynthesis of the siderophore mycobactin and is a potential target for inhibition (Figure 1). The salicylate synthase activity of MbtI produces salicylate and pyruvate from chorismate through an isochorismate intermediate. Inhibition of MbtI activity would decrease the production of salicylate and therefore the synthesis of mycobactin; leading to a decrease in iron acquisition and pathogenesis of ''M. tuberculosis''<ref name= "7a"/> <ref name="1a"/><ref name="5a"/>. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | MbtI catalyses the first committed step in the biosynthesis of the siderophore mycobactin and is a potential target for inhibition. The salicylate synthase activity of MbtI produces salicylate and pyruvate from chorismate through an isochorismate intermediate. Inhibition of MbtI activity would decrease the production of salicylate and therefore the synthesis of mycobactin; leading to a decrease in iron acquisition and pathogenesis of ''M. tuberculosis''<ref name= "7a"/> . | + | |
| - | + | ||
[[Image:Screen Shot 2015-04-10 at 1.27.15 PM.png|500 px|center|thumb|Figure 3: Reaction catalyzed by MbtI in the mycobactin biosynthesis pathway<ref name= "2a"/>.]] | [[Image:Screen Shot 2015-04-10 at 1.27.15 PM.png|500 px|center|thumb|Figure 3: Reaction catalyzed by MbtI in the mycobactin biosynthesis pathway<ref name= "2a"/>.]] | ||
Revision as of 23:39, 26 April 2015
Contents |
Mycobacterium tuberculosis salicylate synthase (Mbt1)
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Ferrer S, Marti S, Moliner V, Tunon I, Bertran J. Understanding the different activities of highly promiscuous MbtI by computational methods. Phys Chem Chem Phys. 2012 Mar 14;14(10):3482-9. doi: 10.1039/c2cp23149b. Epub, 2012 Feb 3. PMID:22307014 doi:http://dx.doi.org/10.1039/c2cp23149b
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Manos-Turvey A, Bulloch EM, Rutledge PJ, Baker EN, Lott JS, Payne RJ. Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI). ChemMedChem. 2010 Jul 5;5(7):1067-79. PMID:20512795 doi:10.1002/cmdc.201000137
- ↑ 3.0 3.1 Lamb AL. Pericyclic reactions catalyzed by chorismate-utilizing enzymes. Biochemistry. 2011 Sep 6;50(35):7476-83. doi: 10.1021/bi2009739. Epub 2011 Aug, 12. PMID:21823653 doi:http://dx.doi.org/10.1021/bi2009739
- ↑ 4.0 4.1 4.2 4.3 De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE 3rd. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci U S A. 2000 Feb 1;97(3):1252-7. PMID:10655517
- ↑ Zwahlen J, Kolappan S, Zhou R, Kisker C, Tonge PJ. Structure and mechanism of MbtI, the salicylate synthase from Mycobacterium tuberculosis. Biochemistry. 2007 Jan 30;46(4):954-64. PMID:17240979 doi:10.1021/bi060852x
- ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 6.11 6.12 6.13 6.14 6.15 6.16 Chi G, Manos-Turvey A, O'Connor PD, Johnston JM, Evans GL, Baker EN, Payne RJ, Lott JS, Bulloch EM. Implications of Binding Mode and Active Site Flexibility for Inhibitor Potency against the Salicylate Synthase from Mycobacterium tuberculosis. Biochemistry. 2012 Jun 7. PMID:22607697 doi:10.1021/bi3002067
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 7.8 Harrison AJ, Yu M, Gardenborg T, Middleditch M, Ramsay RJ, Baker EN, Lott JS. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J Bacteriol. 2006 Sep;188(17):6081-91. PMID:16923875 doi:http://dx.doi.org/188/17/6081
- ↑ 8.0 8.1 8.2 8.3 8.4 Manos-Turvey A, Cergol KM, Salam NK, Bulloch EM, Chi G, Pang A, Britton WJ, West NP, Baker EN, Lott JS, Payne RJ. Synthesis and evaluation of M. tuberculosis salicylate synthase (MbtI) inhibitors designed to probe plasticity in the active site. Org Biomol Chem. 2012 Dec 14;10(46):9223-36. doi: 10.1039/c2ob26736e. Epub 2012, Oct 29. PMID:23108268 doi:http://dx.doi.org/10.1039/c2ob26736e
- ↑ 9.0 9.1 He Z, Stigers Lavoie KD, Bartlett PA, Toney MD. Conservation of mechanism in three chorismate-utilizing enzymes. J Am Chem Soc. 2004 Mar 3;126(8):2378-85. PMID:14982443 doi:http://dx.doi.org/10.1021/ja0389927
- ↑ Tuberculosis (TB). Ed. Sam Posner. Centers for Disease Control and Prevention, n.d. Web. 9 Apr. 2015.
Student contributors
Stephanie Raynor and Robin Gagnon
Related pdb files and proteopedia pages
3D structures of isochorismate pyruvate lyase
3log – MtIPL/isochorismate synthase - Mycobacterium tuberculosis
3rv6, 3rv7, 3rv8, 3rv9, 3st6, 3veh - MtIPL/isochorismate synthase + inhibitor
2h9c – PaIPL residues 1-99 – Pseudomonas aeruginosa
2h9d - PaIPL + pyruvate
3LOG
3D structure of isochorismate synthase
2eua, 3bzm, 3bzn - MenF from E. coli
3os6 - DhbC from Bacillus anthracis
3gse - MenF from Yersinia pestis
3hwo - EntC
3D structure of salicylate synthase
3veh - MbtI with inhibitor methylAMT
3st6 - MbtI with isochorismate analogue inhibitor
3rv6 (Phenyl R-group), 3rv7 (Isopropyl R-group), 3rv8 (Cyclopropyl R-group), 3rv9 (Ethyl R-group) - MbtI with inhibitor
2fn0, 2fn1 (with products salicylate and pyruvate) - Irp9 from Yersinia enterocolitica
2i6y - MbtI