Regulator of G protein signaling
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
<scene name='70/701447/Rgs_monomer/13'>Monomer structure of RGS4</scene> in darkmagenta cartoon diagram: The RGS4 domain corresponds to an array of nine α-helices that fold into two small subdomains. The terminal subdomain contains the N and C termini of the box and is formed by α1, α2, α3, α8, and α9. Helices α1 and α9 lie in antiparallel orientation, juxtaposing the N and C termini of the box. The larger bundle subdomain, formed by α4, α5, α6, and α7, is a classic right-handed, antiparallel four-helix bundle. Both subdomains are required for GAP activity. | <scene name='70/701447/Rgs_monomer/13'>Monomer structure of RGS4</scene> in darkmagenta cartoon diagram: The RGS4 domain corresponds to an array of nine α-helices that fold into two small subdomains. The terminal subdomain contains the N and C termini of the box and is formed by α1, α2, α3, α8, and α9. Helices α1 and α9 lie in antiparallel orientation, juxtaposing the N and C termini of the box. The larger bundle subdomain, formed by α4, α5, α6, and α7, is a classic right-handed, antiparallel four-helix bundle. Both subdomains are required for GAP activity. | ||
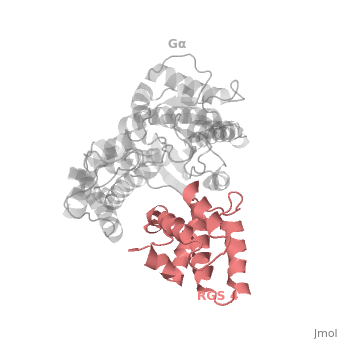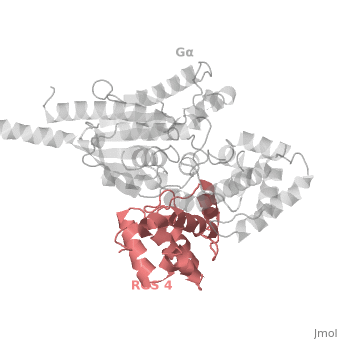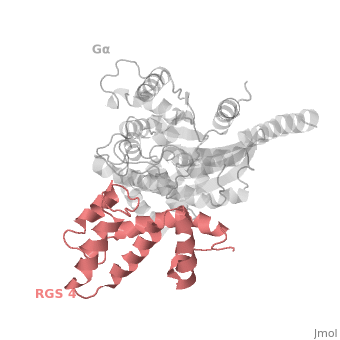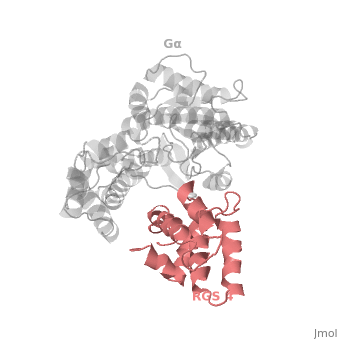
| - | Gα<sub>i1</sub> subunits adopt a conserved fold composed of α helical domain (light gray cartoon), a helical domain of six α helices and a GTPase domain (dark gray cartoon).The GTPase domain hydrolyzes GTP and provides most of Gα's binding surfaces for Gβγ, receptors, effectors and RGS proteins. The GTPase domain contains three flexible regions designated <scene name='70/701447/Gi-s1/6'>switch-I</scene> presented as blue sticks, <scene name='70/701447/Gi-s2/ | + | Gα<sub>i1</sub> subunits adopt a conserved fold composed of α helical domain (light gray cartoon), a helical domain of six α helices and a GTPase domain (dark gray cartoon).The GTPase domain hydrolyzes GTP and provides most of Gα's binding surfaces for Gβγ, receptors, effectors and RGS proteins. The GTPase domain contains three flexible regions designated <scene name='70/701447/Gi-s1/6'>switch-I</scene> presented as blue sticks, <scene name='70/701447/Gi-s2/3'>switch-II</scene> presented as magenta sticks and <scene name='70/701447/Gi-s3/1'>switch-III</scene> presented as green sticks that change conformation in response to GTP binding and hydrolysis. The three switch regions of Gα<sub>i1</sub>: residues 176–184, 201–215, and 233–241, respectively (red cartoon). GDP–Mg<sup>+2</sup>+2, bound in the active site of Gα<sub>i1</sub> is shown as a ball-and-stick model. For clarity, AlF<sub>4</sub> is omitted from the figure.<ref>PMID: 9108480</ref> |
</StructureSection> | </StructureSection> | ||
Revision as of 09:40, 14 May 2015
Regulator of G protein signaling (RGS) interactions with G proteins – RGS4-Gαi as a model structure.
| |||||||||||
References
- ↑ Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY. Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions. Nat Struct Mol Biol. 2011 Jun 19;18(7):846-53. doi: 10.1038/nsmb.2068. PMID:21685921 doi:http://dx.doi.org/10.1038/nsmb.2068
- ↑ Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997 Apr 18;89(2):251-61. PMID:9108480
Proteopedia Page Contributors and Editors (what is this?)
Ali Asli, Denise Salem, Michal Harel, Joel L. Sussman, Jaime Prilusky