Beta2 adrenergic receptor-Gs protein complex
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
GPCRs bind their ligand and [[Group:SMART:A Physical Model of the β2-Adrenergic Receptor|overcome a conformational change]] which allows a [[Guanine nucleotide-binding protein]] (G protein) to detach from the cellular end of the receptor and start the different signal transduction pathways. | GPCRs bind their ligand and [[Group:SMART:A Physical Model of the β2-Adrenergic Receptor|overcome a conformational change]] which allows a [[Guanine nucleotide-binding protein]] (G protein) to detach from the cellular end of the receptor and start the different signal transduction pathways. | ||
Since these receptors have seven transmembrane helices as well as inner and outer cell regions, they are very difficult to purify and crystalize. Some crystal structures have been determined for the inactive receptors as well as for the G protein that they bind. 3sn6 is the first structure of the full complex of the Beta 2 Adrenergic Receptor bound to Gs in their active state, and it provides the first high-resolution insight into the mechanism of signal transduction across the plasma membrane by a GPCR. | Since these receptors have seven transmembrane helices as well as inner and outer cell regions, they are very difficult to purify and crystalize. Some crystal structures have been determined for the inactive receptors as well as for the G protein that they bind. 3sn6 is the first structure of the full complex of the Beta 2 Adrenergic Receptor bound to Gs in their active state, and it provides the first high-resolution insight into the mechanism of signal transduction across the plasma membrane by a GPCR. | ||
| + | |||
| + | == Complex structure == | ||
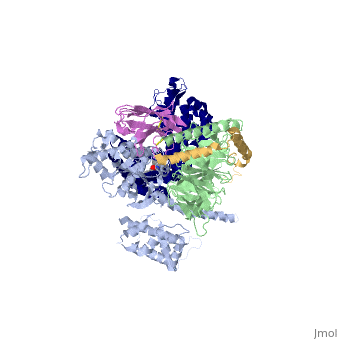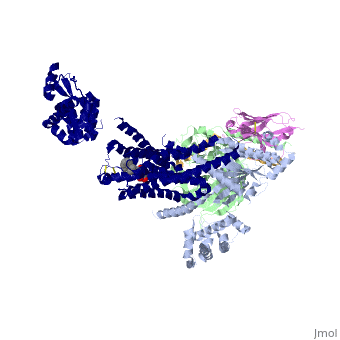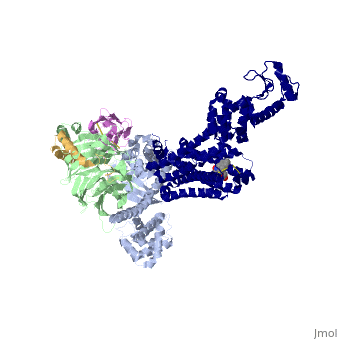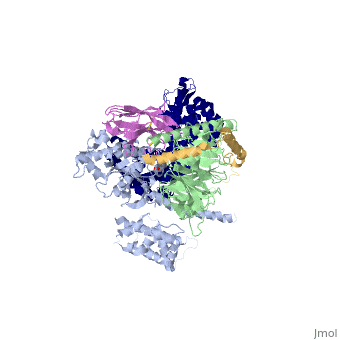
The overall structure shows the b2AR (dark blue) bound to an agonist (in spheres) along with a T4 lysozyme fused to its amino terminus in order to facilitate crystallization. The receptor interacts with Gas (light blue).Gas together with Gb (light green) and Gc (gold) constitute the heterotrimeric G protein Gs. A Gs-binding nanobody which also facilitates crystallization (pink) binds the G protein between the a and b subunits. | The overall structure shows the b2AR (dark blue) bound to an agonist (in spheres) along with a T4 lysozyme fused to its amino terminus in order to facilitate crystallization. The receptor interacts with Gas (light blue).Gas together with Gb (light green) and Gc (gold) constitute the heterotrimeric G protein Gs. A Gs-binding nanobody which also facilitates crystallization (pink) binds the G protein between the a and b subunits. | ||
| - | == G Protein Cycle == | + | == G-Protein-GPCR Intercations == |
| + | The a5-helix of Gas docks into a cavity formed on the intracellular side of the receptor by the opening of transmembrane helices 5 and 6. Within the transmembrane core, the interactions are primarily non-polar. An exception involves <scene name='70/701430/Receptor_g_protein_interaction/1'>packing of Tyr 391 of the a5-helix against Arg 131 of the conservedDRYsequence inTM3.</scene> | ||
| + | |||
| + | == G-Protein Cycle == | ||
[[Image:ImgSmall1.jpg|500px|G protein cycle for the b2AR–Gs complex. Reprinted by permission from Macmillan Publishers Ltd on behalf of Cancer Research UK: Nature 477, 549–555, copyright 2011]] | [[Image:ImgSmall1.jpg|500px|G protein cycle for the b2AR–Gs complex. Reprinted by permission from Macmillan Publishers Ltd on behalf of Cancer Research UK: Nature 477, 549–555, copyright 2011]] | ||
Revision as of 10:04, 3 June 2015
Beta2 adrenergic receptor-Gs protein complex
| |||||||||||
References
- ↑ Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011 Jul 19;477(7366):549-55. doi: 10.1038/nature10361. PMID:21772288 doi:10.1038/nature10361
Proteopedia Page Contributors and Editors (what is this?)
Dan Elran, Michal Harel, Alexander Berchansky, Joel L. Sussman