Sandbox Reserved 1051
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
===Cys 209=== | ===Cys 209=== | ||
| - | <scene name='69/694218/Cysteine_209/2'>Cysteine 209</scene> is located near the Ag85C active site, but is not directly involved in the catalytic mechanism. However, formation of a functional catalytic triad relies on upon a <scene name='69/694220/C209/1'>Van der Waals interaction between C209 and the peptide bond between L232 and T231</scene>. The C209 facilitated interaction stabilizes a <scene name='69/694220/Alpha_9_helix/2'>kinked conformation of the α9 helix</scene> that promotes optimal interaction distances between catalytic residues | + | <scene name='69/694218/Cysteine_209/2'>Cysteine 209</scene> is located near the Ag85C active site, but is not directly involved in the catalytic mechanism. However, formation of a functional catalytic triad relies on upon a <scene name='69/694220/C209/1'>Van der Waals interaction between C209 and the peptide bond between L232 and T231</scene>. The C209 facilitated interaction stabilizes a <scene name='69/694220/Alpha_9_helix/2'>kinked conformation of the α9 helix</scene> that promotes optimal interaction distances between catalytic residues. |
| - | + | ||
| - | == Methods of Inhibition == | ||
| - | + | ==Variants== | |
| + | ===Covalent Modification of Cys209=== | ||
| + | [[Image:Ebselen_inhibition.jpeg|200 px|right|thumb|'''Figure 5:''' Ebselen inhibition relaxing the alpha-9 helix in [http://www.rcsb.org/pdb/explore/explore.do?structureId=4qdu Ag85C-Ebselen]]] | ||
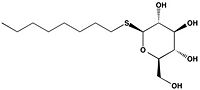
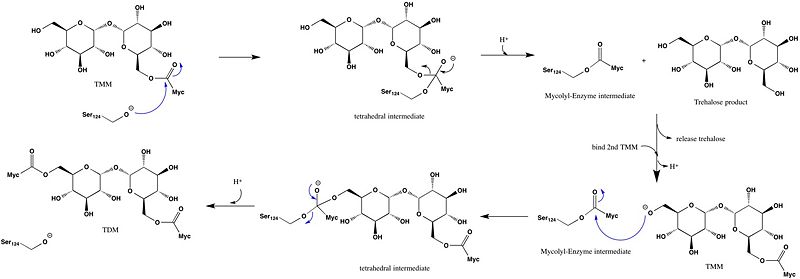
| + | Ag85C can be inhibited by [http://en.wikipedia.org/wiki/Ebselen ebselen], which covalently bounds to the sulfur in C209. Ebselen is a thiol-modifying agent that serves as an electrophile for a C209 nucleophilic attack that results in sulfur-selenium bond formation. Co-crystallization of ebselen with Ag85C provides an explanation for the mechanism of ebselen-based inhibition. The addition of ebselen increases the distance between C209 and L232-T231, which effectively disrupts the interaction that holds the α9 helix in the active conformation. The disruption of this interaction causes the α9 helix to <scene name='69/694220/Inhibited_relaxed_helix/1'>relax</scene>. Furthermore, the bulk of ebselen creates steric hindrance with the α9 helix residues, which can be seen in '''Figure 5'''. The pink helix represents the native enzyme, and the tan helix represents Ag85C covalently bound to ebselen, which is shown in green. Relaxation of the α9 helix due to ebselen removes E228 and H260, which now interacts with S148, from the active site. The absence of these residues destroys the charge relay mechanism, and as a result, the nucleophilicity of the S124 alcohol is not longer strengthened, which decreases serine hydrolytic activity. | ||
| - | [[Image: | + | ===Mutation of Cys209=== |
| + | ====E228Q==== | ||
| + | [[Image:E228Q zoom.png |100 xp|left|thumb|'''Figure 7.''' [http://proteopedia.org/wiki/index.php/4qdz Ag85C-E228Q] active site. The glutamate residue is shifted four angstroms in the mutated form of this enzyme causing a rearrangement of hydrogen bonds within the enzyme. The histidine residue, labeled in pink, takes on two conformations, binding with alternative serine residues, labeled in red.]] | ||
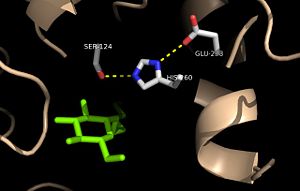
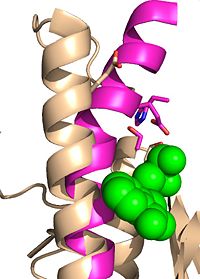
| + | The mutation introduced in [http://proteopedia.org/wiki/index.php/4qdz Ag85C-E228Q]causes the Glu228 of the catalytic triad to be shifted 4 angstroms from its original position in the native structure of Ag85C (Figure 7). Due to the shift of Glu228 is the loss of hydrogen bonds between Ser124 and His260. Instead, <scene name='69/694218/4qdz/2'>His260 bonds with Ser148</scene>, which also results from the shift of Glu228. A weak electron density difference in the His260 position of the native and mutated structures was also noted, suggesting that the residue may take on two alternative conformations in the <scene name='69/694218/Ag85c-e228q/1'>Ag85C-E228Q</scene> mutant. Overall, the enzyme functionality is decreased to only 17% activity.<ref name="Favrot"/> | ||
| - | Ag85C | + | Unlike other Ag85C mutants and modifications aforementioned, the <scene name='69/694218/Ag85c-e228q/1'>Ag85C-E228Q</scene>does not eliminate any hydrogen bonds in the catalytic triad. Rather, it simply replaces a carboxylate moiety with an amide. Further, the structural change observed in the low-energy conformation of the <scene name='69/694218/Ag85c-e228q/1'>Ag85C-E228Q</scene> mutant provides additional support for the hypothesis that the natively kinked helix α-9 is central to the enzymatic function of Ag85C.<ref name="Favrot"/> |
| + | |||
| + | {{clear}} | ||
| + | ====H260Q==== | ||
| - | + | [[Image:H260Q zoom.png|100 xp|left|thumb|'''Figure 8.''' [http://proteopedia.org/wiki/index.php/4qe3 Ag85C-H260Q] active site. A shift in helix α-9 prevents formation of any stabilizing hydrogen bonds between residues His260 and Glu228, thus decreasing its enzymatic activity.]] | |
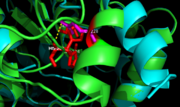
| + | In <scene name='69/694218/Mutationh260q/1'>Ag85C-H260Q</scene>, a shift in helix α-9 <scene name='69/694218/Ag85c-hg/1'>prevents the hydrogen bond formation</scene>between residues His260 and Glu228, thus decreasing its enzymatic activity (Figure 8). The conversion of the glutamate, a key player in the catalytic triad, to the corresponding amide-containing side chain as well as a loss of a general base in the charge relay are both key causes for the loss of function.<ref name="Favrot"/> | ||
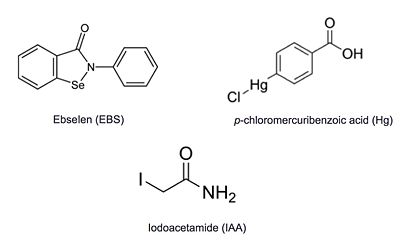
Additional thiol-modifying agents, [http://en.wikipedia.org/wiki/4-Chloromercuribenzoic_acid p-chloromercuribenzoic acid] and [http://en.wikipedia.org/wiki/Iodoacetamide iodoacetamide], were crystalized with Ag85C. The structures show that each of these thiol-reactive inhibitors covalently bound to C209 and caused a relaxation of the α9 helix in a similar fashion to ebselen. | Additional thiol-modifying agents, [http://en.wikipedia.org/wiki/4-Chloromercuribenzoic_acid p-chloromercuribenzoic acid] and [http://en.wikipedia.org/wiki/Iodoacetamide iodoacetamide], were crystalized with Ag85C. The structures show that each of these thiol-reactive inhibitors covalently bound to C209 and caused a relaxation of the α9 helix in a similar fashion to ebselen. | ||
Revision as of 17:33, 15 June 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
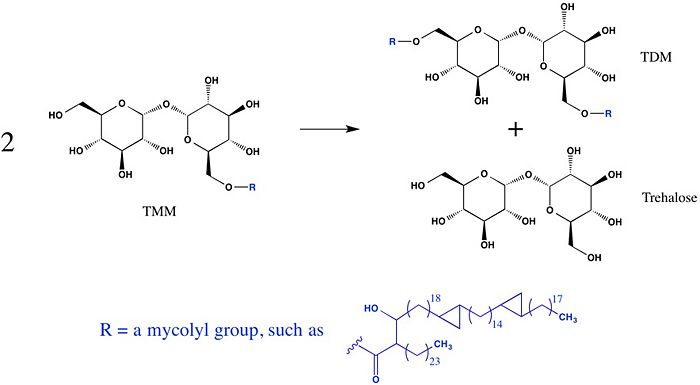
Trehalose-O-mycolyltransferase Ag85C
Introduction
Antigen 85C is one of three homologous protein components of the Ag85 complex in the cell wall of M. tuberculosis. This serine esterase enzyme catalyzes the transfer of mycolyl groups, characteristic components of the cell wall of mycobacteria. Several three dimensional structures of Ag85C have been solved, including the wild type enzyme as well as active site variants due to site-directed mutagenesis and covalent modification.
| |||||||||||
References
- ↑ Jackson M, Raynaud C, Laneelle MA, Guilhot C, Laurent-Winter C, Ensergueix D, Gicquel B, Daffe M. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol Microbiol. 1999 Mar;31(5):1573-87. PMID:10200974
- ↑ Ronning DR, Klabunde T, Besra GS, Vissa VD, Belisle JT, Sacchettini JC. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat Struct Biol. 2000 Feb;7(2):141-6. PMID:10655617 doi:10.1038/72413
- ↑ Ronning DR, Vissa V, Besra GS, Belisle JT, Sacchettini JC. Mycobacterium tuberculosis antigen 85A and 85C structures confirm binding orientation and conserved substrate specificity. J Biol Chem. 2004 Aug 27;279(35):36771-7. Epub 2004 Jun 10. PMID:15192106 doi:http://dx.doi.org/10.1074/jbc.M400811200
- ↑ Favrot L, Lajiness DH, Ronning DR. Inactivation of the Mycobacterium tuberculosis Antigen 85 complex by covalent, allosteric inhibitors. J Biol Chem. 2014 Jul 14. pii: jbc.M114.582445. PMID:25028518 doi:http://dx.doi.org/10.1074/jbc.M114.582445
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedFavrot