Sandbox Reserved 1076
From Proteopedia
| Line 11: | Line 11: | ||
== General Structure and Function == | == General Structure and Function == | ||
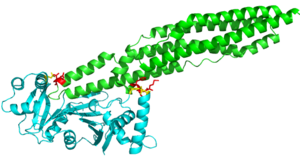
| - | The EspG3 protein shown in Figure 1 has a mass of 33.7kD <ref>PMID:25275011</ref>. The proteins beta sheets make up the | + | The EspG3 protein shown in Figure 1 has a mass of 33.7kD <ref>PMID:25275011</ref>. The proteins beta sheets make up the core of the protein (yellow). As a monomeric protein, this binds to its ligand with high specificity. A key beta sheet region on the <scene name='69/694242/Espg3_differences/4'>β-2 and β-3 strands</scene> will vary between EspG subtypes to influence what the random loop on the PE-PPE protein will interact with, since this region includes key residues that interact with the random coil of the [http://proteopedia.org/wiki/index.php/PE/PPE_Protein_Complex PE-PPE] ligand. The β-sheet core is surrounded by 8 alpha helices (red) which add to the structure of the protein. One key <scene name='69/694242/Espg3_differences/2'>alpha helix</scene> will vary per EspG protein to stericly limit binding to PE-PPE ligand. The random coil (green) connects the beta sheets and helices together, where the long <scene name='69/694242/Espg3_differences/3'>random loop</scene> variations can also impact binding to the EspG's ligand. |
| + | |||
Through specific binding factors, an EspG binds to its PE-PPE ligand to be secreted through the ESAT pathway. Though the ESAT-6 secretion system is poorly understood, it is known that PE-PPE proteins and EspG proteins influence virulence and pathogenicity of the infection. | Through specific binding factors, an EspG binds to its PE-PPE ligand to be secreted through the ESAT pathway. Though the ESAT-6 secretion system is poorly understood, it is known that PE-PPE proteins and EspG proteins influence virulence and pathogenicity of the infection. | ||
| + | ==Structural Differences between EspG subtypes== | ||
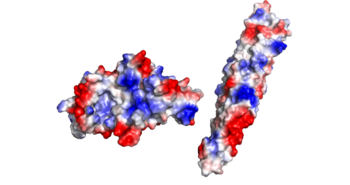
| - | + | [[Image:EspG5_and_EspG3_overlap_W.png|250 px|left|thumb|Figure 2: EspG5 (blue) and EspG3 (yellow)]] | |
| - | + | ||
| - | [[Image:EspG5_and_EspG3_overlap_W.png|250 px|left|thumb|EspG5 and EspG3]] | + | |
| - | + | Featured in Figure 2 are the key differences found on the tertiary structure of EspG3 (yellow) with EspG5 (blue). The highlighted alpha helix (lower right) shows a difference in length between EspG3 and EspG5. This difference in length contributes to the steric hinderance when binding to a PE-PPE ligand. The random loop highlighted toward the bottom of this protein also varies in length between EspG proteins, and this influences ligand binding. The small random turn highlighted shows inconceivable difference to the EspG5 protein. The sequence similarity between EspG5 and EspG3 at the β-2 and β-3 strands is very low. | |
Revision as of 22:45, 15 June 2015
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
EspG5 Secretion Protein
Introduction
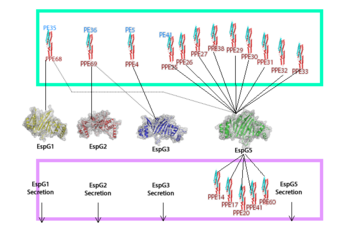
EspG is a key secretion protein involved with the virulence of Mycobacterium tuberculosis. The specificity of EspG binding affinity to its specific PE-PPE ligand has many contributing factors. The four different EspG proteins found in Mycobacterium tuberculosis have different characteristics that influence binding, where EspG5 binds to the most PE-PPE proteins. Not all EspG proteins bind to the same ligand; specific interactions from specific residue interactions, electrostatics, steric hinderance and concavity of the EspG binding pocket influence binding. The EspG PE-PPE complex is to be excreted in the ESAT-6 pathway, this pathway is an attractive target for inducing apoptosis in Mtb, this makes it a good drug target.
| |||||||||||
References
- ↑ Ekiert DC, Cox JS. Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14758-63. doi:, 10.1073/pnas.1409345111. Epub 2014 Oct 1. PMID:25275011 doi:http://dx.doi.org/10.1073/pnas.1409345111
- ↑ Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, Frenkiel TA, Gordon SV, Hewinson RG, Burke B, Norman J, Williamson RA, Carr MD. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 2005 Jul 20;24(14):2491-8. Epub 2005 Jun 23. PMID:15973432
Similar Pages
Student Contributors
- Mark Meredith
- Jonathan Golliher