Group:MUZIC:actinin2
From Proteopedia
(Difference between revisions)

| Line 4: | Line 4: | ||
Muscle cells are responsible for the voluntary and involuntary contraction. These cells contains myofibrils, which are bundles mainly formed by actin and myosin filaments. These filaments are organized into repetitive subunits, called sarcomeres, which are connected in tandem by Z-disks, constituting an intricate macromolecular assembly. A plethora of proteins have been identified in the Z-disk [[http://www.proteopedia.org/wiki/index.php/Group:MUZIC:Interactome]], members of different protein classes - globular, intrinsically disordered and multi-domain proteins, but the details about their structure and interaction network at molecular level are still an enigma. Understanding how these proteins work together and how they interact with other molecules can have major impacts in medicine. | Muscle cells are responsible for the voluntary and involuntary contraction. These cells contains myofibrils, which are bundles mainly formed by actin and myosin filaments. These filaments are organized into repetitive subunits, called sarcomeres, which are connected in tandem by Z-disks, constituting an intricate macromolecular assembly. A plethora of proteins have been identified in the Z-disk [[http://www.proteopedia.org/wiki/index.php/Group:MUZIC:Interactome]], members of different protein classes - globular, intrinsically disordered and multi-domain proteins, but the details about their structure and interaction network at molecular level are still an enigma. Understanding how these proteins work together and how they interact with other molecules can have major impacts in medicine. | ||
| - | The protein α-actinin skeletal muscle isoform 2 (α-actinin-2) has a pivotal role in the formation and integrity of the ultra-structure of striated muscle Z-disk. Including, in the beginning steps of progression of the [[Z-bodies]] in premyofibrils and nascent myofibrils to Z-disks of the mature [[myofibrils]] <ref>PMID: 16465476</ref>. | + | The protein α-actinin skeletal muscle isoform 2 (α-actinin-2) has a pivotal role in the formation and integrity of the ultra-structure of striated muscle Z-disk<ref>PMID: 25433700</ref>. Including, in the beginning steps of progression of the [[Z-bodies]] in premyofibrils and nascent myofibrils to Z-disks of the mature [[myofibrils]] <ref>PMID: 16465476</ref>. |
α-actinin-2 is a calcium-independent isoform expressed in all muscle fibers. The expression in human skeletal muscle overlaps α-actinin-3. Both proteins form heterodimers in vitro and in vivo and share high protein sequence identity (80%), therefore suggesting a high 3-D structure similarity. They are regulated by phosphoinositides, rendering alpha-actinin capable of binding to titin (and some other Z-disk partners). It is worth pointing out that the other α-actinin non-muscles isoforms (1 and 4) are calcium-dependent. α-actinin-1 is associate with cell adhesion molecules, stabilizing cell adhesion and regulating cell shape and cell motility, while α-actinin-4 are related to the cytoskeleton playing an essential role in cell motility and shape and tumor suppressor activity. | α-actinin-2 is a calcium-independent isoform expressed in all muscle fibers. The expression in human skeletal muscle overlaps α-actinin-3. Both proteins form heterodimers in vitro and in vivo and share high protein sequence identity (80%), therefore suggesting a high 3-D structure similarity. They are regulated by phosphoinositides, rendering alpha-actinin capable of binding to titin (and some other Z-disk partners). It is worth pointing out that the other α-actinin non-muscles isoforms (1 and 4) are calcium-dependent. α-actinin-1 is associate with cell adhesion molecules, stabilizing cell adhesion and regulating cell shape and cell motility, while α-actinin-4 are related to the cytoskeleton playing an essential role in cell motility and shape and tumor suppressor activity. | ||
| Line 24: | Line 24: | ||
== Structures == | == Structures == | ||
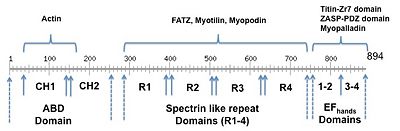
| - | + | Current structural data of the isoform 2 suggest an autoinhibited closed conformation where the C-terminal lobe of the calmodulin-like domain is bound to the α-helix that connects the ABD domain to the first spectrin-like repeat. A gallery of additional structures of α-actinin fragments are also available (see the link)[[http://www.proteopedia.org/wiki/index.php/Actinin#3D_Structures_of_Actinin]]. The ABD domain reveals the arrangement of the two calponin homology domains (CH1 and CH2) in a closed conformation<ref name="ABD domain"/> | |
| - | The | + | The rod domain shows the four spectrins like domain of one chain forms a homo anti-parallel dimer, which is a left handed twisted from one end of the rod to the other end <ref>PMID:11470434</ref>. |
The solution structure of the complex between the human α-actinin-2 EF-hand domain and the chicken titin Z-repeat 7 domain was solved by NMR spectroscopy. A specific set of contacts and some important residues for protein-protein interactions were identified. It was shown that recognition and positioning of Z-repeat in cavity of EF-hand domain is mediated by balance of electrostatic and hydrophobic interactions. The reported semi-open conformation of complex is commonly seen in homologous structures. This conformation seems to be typical for calcium-independent recognition by EF-hand domain and is fully compatible with formation of stable long-term complexes between titin and α-actinin-2. | The solution structure of the complex between the human α-actinin-2 EF-hand domain and the chicken titin Z-repeat 7 domain was solved by NMR spectroscopy. A specific set of contacts and some important residues for protein-protein interactions were identified. It was shown that recognition and positioning of Z-repeat in cavity of EF-hand domain is mediated by balance of electrostatic and hydrophobic interactions. The reported semi-open conformation of complex is commonly seen in homologous structures. This conformation seems to be typical for calcium-independent recognition by EF-hand domain and is fully compatible with formation of stable long-term complexes between titin and α-actinin-2. | ||
Current revision
| |||||||||||
References
- ↑ Ribeiro Ede A Jr, Pinotsis N, Ghisleni A, Salmazo A, Konarev PV, Kostan J, Sjoblom B, Schreiner C, Polyansky AA, Gkougkoulia EA, Holt MR, Aachmann FL, Zagrovic B, Bordignon E, Pirker KF, Svergun DI, Gautel M, Djinovic-Carugo K. The Structure and Regulation of Human Muscle alpha-Actinin. Cell. 2014 Dec 4;159(6):1447-60. doi: 10.1016/j.cell.2014.10.056. Epub 2014 Nov, 26. PMID:25433700 doi:http://dx.doi.org/10.1016/j.cell.2014.10.056
- ↑ Sanger JW, Kang S, Siebrands CC, Freeman N, Du A, Wang J, Stout AL, Sanger JM. How to build a myofibril. J Muscle Res Cell Motil. 2005;26(6-8):343-54. PMID:16465476 doi:10.1007/s10974-005-9016-7
- ↑ 3.0 3.1 Franzot G, Sjoblom B, Gautel M, Djinovic Carugo K. The crystal structure of the actin binding domain from alpha-actinin in its closed conformation: structural insight into phospholipid regulation of alpha-actinin. J Mol Biol. 2005 Apr 22;348(1):151-65. PMID:15808860 doi:http://dx.doi.org/10.1016/j.jmb.2005.01.002
- ↑ Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, Valle G, Lanfranchi G. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 1999 Jul 26;146(2):465-75. PMID:10427098
- ↑ Au Y, Atkinson RA, Guerrini R, Kelly G, Joseph C, Martin SR, Muskett FW, Pallavicini A, Faulkner G, Pastore A. Solution structure of ZASP PDZ domain; implications for sarcomere ultrastructure and enigma family redundancy. Structure. 2004 Apr;12(4):611-22. PMID:15062084 doi:10.1016/j.str.2004.02.019
- ↑ Fukami K, Sawada N, Endo T, Takenawa T. Identification of a phosphatidylinositol 4,5-bisphosphate-binding site in chicken skeletal muscle alpha-actinin. J Biol Chem. 1996 Feb 2;271(5):2646-50. PMID:8576235
- ↑ Fukami K, Furuhashi K, Inagaki M, Endo T, Hatano S, Takenawa T. Requirement of phosphatidylinositol 4,5-bisphosphate for alpha-actinin function. Nature. 1992 Sep 10;359(6391):150-2. PMID:1326084 doi:http://dx.doi.org/10.1038/359150a0
- ↑ Young P, Gautel M. The interaction of titin and alpha-actinin is controlled by a phospholipid-regulated intramolecular pseudoligand mechanism. EMBO J. 2000 Dec 1;19(23):6331-40. PMID:11101506 doi:10.1093/emboj/19.23.6331
- ↑ Ylanne J, Scheffzek K, Young P, Saraste M. Crystal structure of the alpha-actinin rod reveals an extensive torsional twist. Structure. 2001 Jul 3;9(7):597-604. PMID:11470434
- ↑ Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995 Aug 15;92(4):785-9. PMID:7641357
- ↑ Chiu C, Bagnall RD, Ingles J, Yeates L, Kennerson M, Donald JA, Jormakka M, Lind JM, Semsarian C. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J Am Coll Cardiol. 2010 Mar 16;55(11):1127-35. doi: 10.1016/j.jacc.2009.11.016. PMID:20022194 doi:10.1016/j.jacc.2009.11.016
Proteopedia Page Contributors and Editors (what is this?)
Euripides Ribeiro, Alexander Berchansky, Nikos Pinotsis, Michal Harel, Jaime Prilusky