Regulator of G protein signaling
From Proteopedia
(Difference between revisions)
| Line 36: | Line 36: | ||
Many RGS protein residues located in the vicinity of the <scene name='70/701447/Rgs4-ga_interface/3'>RGS domain–Gα interface</scene> (RGS protein shown as wheat cartoon and Gα<sub>i1</sub> subunit shown as white surface) contribute to RGS-G proteins interaction. Based on energy calculation and experimental validation of RGS-Gα complexes from Gα<sub>i</sub> subfamily members, these residues classified into two major groups: <scene name='70/701447/Rgs4-ga-sandc-residues/4'>Significant and Conserved residues</scene> shown as red spheres that are located mainly in the center of the RGS domain–Gα interface and have a primary role in accelerating Gα GTPase by stabilizing Gα in an optimal conformation for GTP hydrolysis. Whereas the <scene name='70/701447/Rgs4-ga_modulatory_residues/1'>putative Modulatory residues</scene> shown as purple spheres are located mostly at the periphery of the interface where they contribute to Gα subunit's recognition.<ref>PMID: 21685921</ref> | Many RGS protein residues located in the vicinity of the <scene name='70/701447/Rgs4-ga_interface/3'>RGS domain–Gα interface</scene> (RGS protein shown as wheat cartoon and Gα<sub>i1</sub> subunit shown as white surface) contribute to RGS-G proteins interaction. Based on energy calculation and experimental validation of RGS-Gα complexes from Gα<sub>i</sub> subfamily members, these residues classified into two major groups: <scene name='70/701447/Rgs4-ga-sandc-residues/4'>Significant and Conserved residues</scene> shown as red spheres that are located mainly in the center of the RGS domain–Gα interface and have a primary role in accelerating Gα GTPase by stabilizing Gα in an optimal conformation for GTP hydrolysis. Whereas the <scene name='70/701447/Rgs4-ga_modulatory_residues/1'>putative Modulatory residues</scene> shown as purple spheres are located mostly at the periphery of the interface where they contribute to Gα subunit's recognition.<ref>PMID: 21685921</ref> | ||
| - | Gα subunits participate in a range of interactions with a variety of other proteins. Therefore, they have interfaces that interact selectively with receptors, effector subfamilies and RGS proteins. However, <scene name='70/701447/Gi-rgs4_interface/4'>Gα residues</scene> from Gα<sub>i</sub> subfamily members that interact specifically with RGS proteins are highly conserved (red spheres). These Gα Residues located on Gα switch regions interact with Significant & Conserved RGS residues because of the pivotal role of the switch regions in GTP hydrolysis that is catalyzed by RGS proteins. On the other hand, Gα residues located in switch regions II and III and multiple residues in the Gα all-helical domain interact with Modulatory RGS residues. For example, one important RGS4 residue that projects into the active site of Gα<sub>i</sub> is r-Asn-128, which contacts the side chains of a- | + | Gα subunits participate in a range of interactions with a variety of other proteins. Therefore, they have interfaces that interact selectively with receptors, effector subfamilies and RGS proteins. However, <scene name='70/701447/Gi-rgs4_interface/4'>Gα residues</scene> from Gα<sub>i</sub> subfamily members that interact specifically with RGS proteins are highly conserved (red spheres). These Gα Residues located on Gα switch regions interact with Significant & Conserved RGS residues because of the pivotal role of the switch regions in GTP hydrolysis that is catalyzed by RGS proteins. On the other hand, Gα residues located in switch regions II and III and multiple residues in the Gα all-helical domain interact with Modulatory RGS residues. For example, one important RGS4 residue that projects into the active site of Gα<sub>i</sub> is r-Asn-128, which contacts the side chains of a-Lys-180, a-Gln-204, and a-Glu-207.<ref>PMID: 21685921</ref> |
</StructureSection> | </StructureSection> | ||
Revision as of 04:56, 14 July 2015
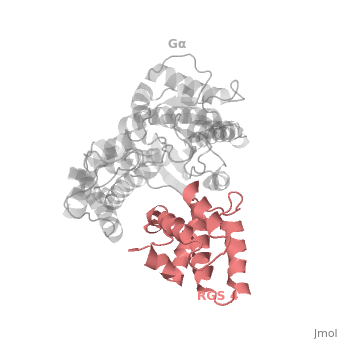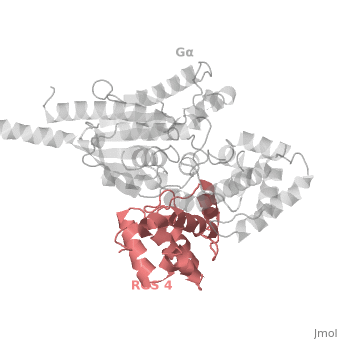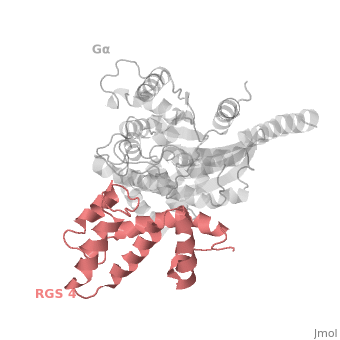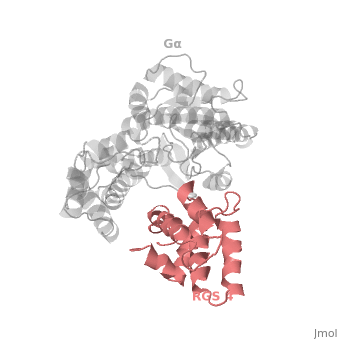
Regulator of G protein signaling (RGS) interactions with G proteins – RGS4-Gαi as a model structure.
| |||||||||||
References
- ↑ Milligan G, Kostenis E. Heterotrimeric G-proteins: a short history. Br J Pharmacol. 2006 Jan;147 Suppl 1:S46-55. PMID:16402120 doi:http://dx.doi.org/10.1038/sj.bjp.0706405
- ↑ Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY. Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions. Nat Struct Mol Biol. 2011 Jun 19;18(7):846-53. doi: 10.1038/nsmb.2068. PMID:21685921 doi:http://dx.doi.org/10.1038/nsmb.2068
- ↑ Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997 Apr 18;89(2):251-61. PMID:9108480
- ↑ Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY. Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions. Nat Struct Mol Biol. 2011 Jun 19;18(7):846-53. doi: 10.1038/nsmb.2068. PMID:21685921 doi:http://dx.doi.org/10.1038/nsmb.2068
- ↑ Kosloff M, Travis AM, Bosch DE, Siderovski DP, Arshavsky VY. Integrating energy calculations with functional assays to decipher the specificity of G protein-RGS protein interactions. Nat Struct Mol Biol. 2011 Jun 19;18(7):846-53. doi: 10.1038/nsmb.2068. PMID:21685921 doi:http://dx.doi.org/10.1038/nsmb.2068
Proteopedia Page Contributors and Editors (what is this?)
Ali Asli, Denise Salem, Michal Harel, Joel L. Sussman, Jaime Prilusky