We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
FMDV 3C
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
FMDV 3C Protease | FMDV 3C Protease | ||
| - | By, Erica Martel | ||
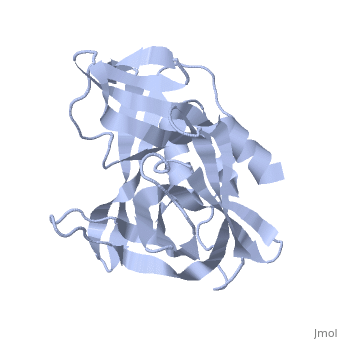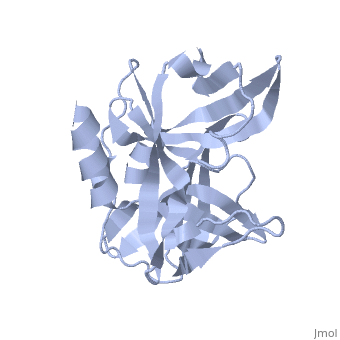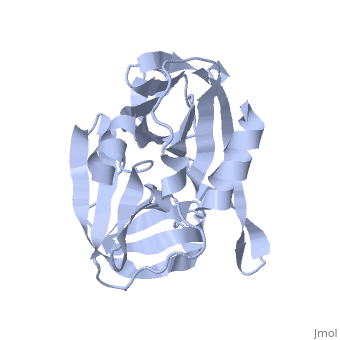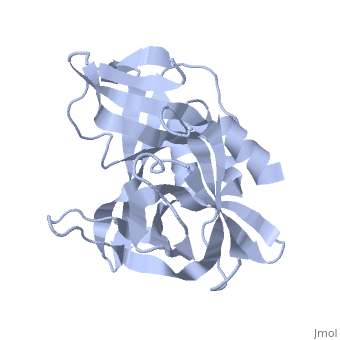
<StructureSection load='2j92' size='340' side='right' caption='FMDV 3C Protease Type A10' scene=''> | <StructureSection load='2j92' size='340' side='right' caption='FMDV 3C Protease Type A10' scene=''> | ||
'''Foot and Mouth Disease 3C Protease'''. | '''Foot and Mouth Disease 3C Protease'''. | ||
| + | '''By, Erica Martel'''. | ||
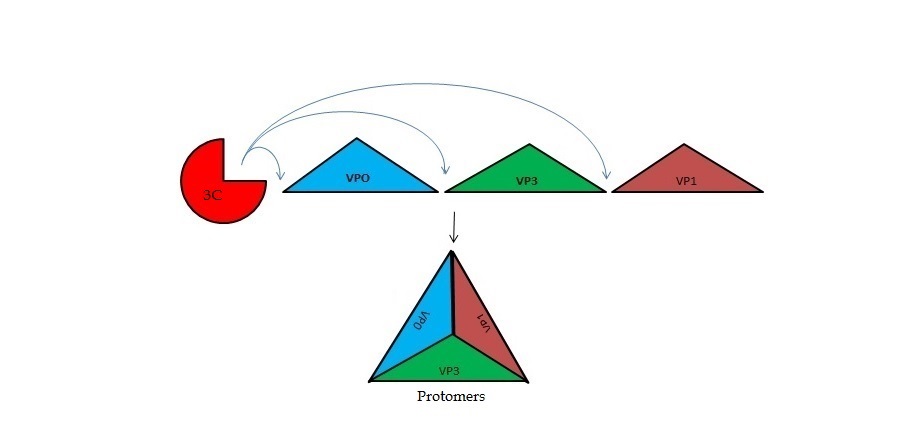
The viral RNA genome is translated as a single polypeptide precursor that must be cleaved into functional proteins by virally encoded proteases. The FMDV 3C protease is responsible for most cleavage in the viral polyprotein (10 out of the 13 cleavages) and has a conserved amino acid sequence across different viral strains, making the enzyme an attractive target for inhibitor design and antiviral drugs. The 3C protease cuts itself out of the polyprotein and is responsible for processing the P1 peptide (with exception of VP4-VP21 (1AB)) as well as P2 and P3, which has lead to its categorization as a catalytic triad. The kinetics of processing proceeds to cleave the bonds in this order of preference: VP3-VP1, VP0-VP3, and VP1-2A, where they then assemble into a triangular protomer (see figure below). Five protomers come together to make pentamers, twelve of which conjoin to complete the virus shell. | The viral RNA genome is translated as a single polypeptide precursor that must be cleaved into functional proteins by virally encoded proteases. The FMDV 3C protease is responsible for most cleavage in the viral polyprotein (10 out of the 13 cleavages) and has a conserved amino acid sequence across different viral strains, making the enzyme an attractive target for inhibitor design and antiviral drugs. The 3C protease cuts itself out of the polyprotein and is responsible for processing the P1 peptide (with exception of VP4-VP21 (1AB)) as well as P2 and P3, which has lead to its categorization as a catalytic triad. The kinetics of processing proceeds to cleave the bonds in this order of preference: VP3-VP1, VP0-VP3, and VP1-2A, where they then assemble into a triangular protomer (see figure below). Five protomers come together to make pentamers, twelve of which conjoin to complete the virus shell. | ||
[[Image:FMDV_3C_Prot.jpeg]] | [[Image:FMDV_3C_Prot.jpeg]] | ||
Revision as of 14:10, 15 July 2015
FMDV 3C Protease
| |||||||||||
References
references [1]