4ald
From Proteopedia
| Line 4: | Line 4: | ||
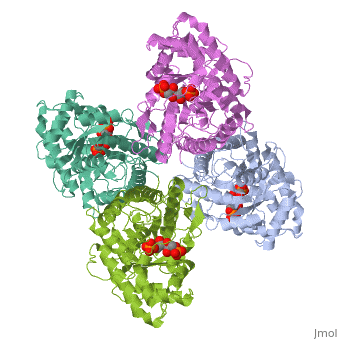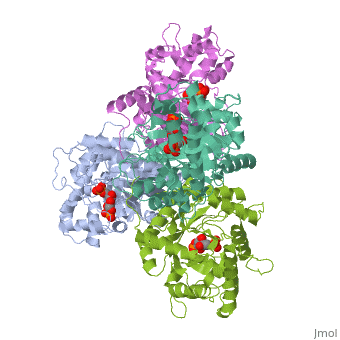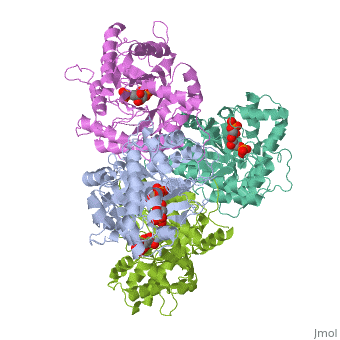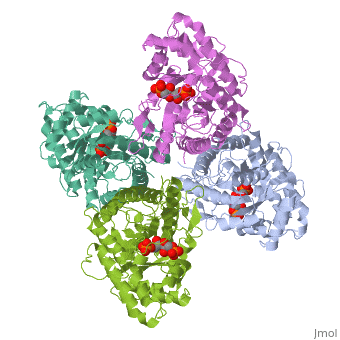
|PDB= 4ald |SIZE=350|CAPTION= <scene name='initialview01'>4ald</scene>, resolution 2.8Å | |PDB= 4ald |SIZE=350|CAPTION= <scene name='initialview01'>4ald</scene>, resolution 2.8Å | ||
|SITE= <scene name='pdbsite=SBL:Schiff'S+Base+LYS'>SBL</scene> | |SITE= <scene name='pdbsite=SBL:Schiff'S+Base+LYS'>SBL</scene> | ||
| - | |LIGAND= <scene name='pdbligand=2FP:1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM)'>2FP</scene> | + | |LIGAND= <scene name='pdbligand=2FP:1,6-FRUCTOSE+DIPHOSPHATE+(LINEAR+FORM)'>2FP</scene> |
| - | |ACTIVITY= [http://en.wikipedia.org/wiki/Fructose-bisphosphate_aldolase Fructose-bisphosphate aldolase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=4.1.2.13 4.1.2.13] | + | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/Fructose-bisphosphate_aldolase Fructose-bisphosphate aldolase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=4.1.2.13 4.1.2.13] </span> |
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4ald FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4ald OCA], [http://www.ebi.ac.uk/pdbsum/4ald PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=4ald RCSB]</span> | ||
}} | }} | ||
| Line 14: | Line 17: | ||
==Overview== | ==Overview== | ||
Fructose 1,6-bisphosphate aldolase catalyzes the reversible cleavage of fructose 1,6-bisphosphate and fructose 1-phosphate to dihydroxyacetone phosphate and either glyceraldehyde 3-phosphate or glyceraldehyde, respectively. Catalysis involves the formation of a Schiff's base intermediate formed at the epsilon-amino group of Lys229. The existing apo-enzyme structure was refined using the crystallographic free-R-factor and maximum likelihood methods that have been shown to give improved structural results that are less subject to model bias. Crystals were also soaked with the natural substrate (fructose 1,6-bisphosphate), and the crystal structure of this complex has been determined to 2.8 A. The apo structure differs from the previous Brookhaven-deposited structure (1ald) in the flexible C-terminal region. This is also the region where the native and complex structures exhibit differences. The conformational changes between native and complex structure are not large, but the observed complex does not involve the full formation of the Schiff's base intermediate, and suggests a preliminary hydrogen-bonded Michaelis complex before the formation of the covalent complex. | Fructose 1,6-bisphosphate aldolase catalyzes the reversible cleavage of fructose 1,6-bisphosphate and fructose 1-phosphate to dihydroxyacetone phosphate and either glyceraldehyde 3-phosphate or glyceraldehyde, respectively. Catalysis involves the formation of a Schiff's base intermediate formed at the epsilon-amino group of Lys229. The existing apo-enzyme structure was refined using the crystallographic free-R-factor and maximum likelihood methods that have been shown to give improved structural results that are less subject to model bias. Crystals were also soaked with the natural substrate (fructose 1,6-bisphosphate), and the crystal structure of this complex has been determined to 2.8 A. The apo structure differs from the previous Brookhaven-deposited structure (1ald) in the flexible C-terminal region. This is also the region where the native and complex structures exhibit differences. The conformational changes between native and complex structure are not large, but the observed complex does not involve the full formation of the Schiff's base intermediate, and suggests a preliminary hydrogen-bonded Michaelis complex before the formation of the covalent complex. | ||
| - | |||
| - | ==Disease== | ||
| - | Known disease associated with this structure: Aldolase A deficiency OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=103850 103850]] | ||
==About this Structure== | ==About this Structure== | ||
| Line 30: | Line 30: | ||
[[Category: Dauter, Z.]] | [[Category: Dauter, Z.]] | ||
[[Category: Littlechild, J A.]] | [[Category: Littlechild, J A.]] | ||
| - | [[Category: 2FP]] | ||
[[Category: complex]] | [[Category: complex]] | ||
[[Category: lyase (aldehyde)]] | [[Category: lyase (aldehyde)]] | ||
| Line 36: | Line 35: | ||
[[Category: type 1 aldolase]] | [[Category: type 1 aldolase]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 05:37:55 2008'' |
Revision as of 02:37, 31 March 2008
| |||||||
| , resolution 2.8Å | |||||||
|---|---|---|---|---|---|---|---|
| Sites: | |||||||
| Ligands: | |||||||
| Activity: | Fructose-bisphosphate aldolase, with EC number 4.1.2.13 | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
HUMAN MUSCLE FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE COMPLEXED WITH FRUCTOSE 1,6-BISPHOSPHATE
Overview
Fructose 1,6-bisphosphate aldolase catalyzes the reversible cleavage of fructose 1,6-bisphosphate and fructose 1-phosphate to dihydroxyacetone phosphate and either glyceraldehyde 3-phosphate or glyceraldehyde, respectively. Catalysis involves the formation of a Schiff's base intermediate formed at the epsilon-amino group of Lys229. The existing apo-enzyme structure was refined using the crystallographic free-R-factor and maximum likelihood methods that have been shown to give improved structural results that are less subject to model bias. Crystals were also soaked with the natural substrate (fructose 1,6-bisphosphate), and the crystal structure of this complex has been determined to 2.8 A. The apo structure differs from the previous Brookhaven-deposited structure (1ald) in the flexible C-terminal region. This is also the region where the native and complex structures exhibit differences. The conformational changes between native and complex structure are not large, but the observed complex does not involve the full formation of the Schiff's base intermediate, and suggests a preliminary hydrogen-bonded Michaelis complex before the formation of the covalent complex.
About this Structure
4ALD is a Single protein structure of sequence from Homo sapiens. The following page contains interesting information on the relation of 4ALD with [The Glycolytic Enzymes]. Full crystallographic information is available from OCA.
Reference
Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications., Dalby A, Dauter Z, Littlechild JA, Protein Sci. 1999 Feb;8(2):291-7. PMID:10048322
Page seeded by OCA on Mon Mar 31 05:37:55 2008