We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
NADP-dependent malic enzyme
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | == | + | == NADP-dependent malic enzyme == |
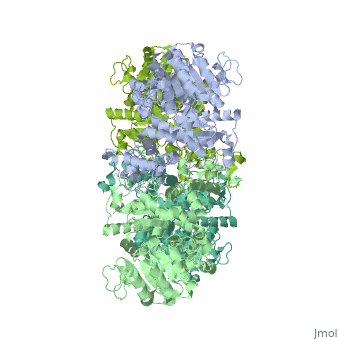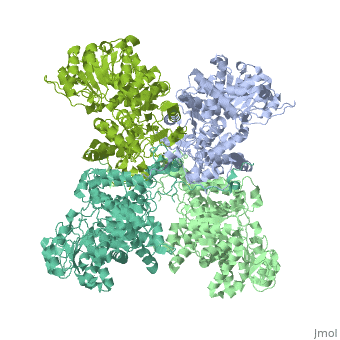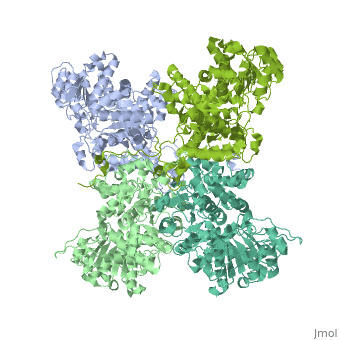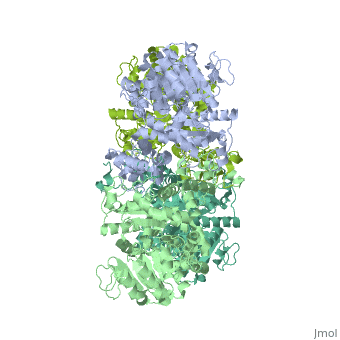
| - | <StructureSection load=' | + | <StructureSection load='3wja' size='340' side='right' caption='The best structure for NADP-dependent malic enzyme shown: 3wja' scene=''> |
| - | Best example is | + | Best example is 3wja and is shown in the viewer.<br> |
| - | Molecule | + | Molecule NADP-dependent malic enzyme, also known as NADP-ME, NADP-dependent malic enzyme and Malic enzyme 1. |
| - | + | ||
| - | + | ||
==Catalytic Activity == | ==Catalytic Activity == | ||
| - | Hydrolysis of (1->4)-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in a peptidoglycan and between N-acetyl-D-glucosamine residues in chitodextrins.Data source: Uniprot [http://www.uniprot.org/uniprot/P00720 P00720]<br> | ||
== Disease == | == Disease == | ||
| Line 15: | Line 12: | ||
== Biological process == | == Biological process == | ||
Is involved in the following biological processes:<br> | Is involved in the following biological processes:<br> | ||
| - | + | malate metabolic process<br> | |
| - | + | oxidation-reduction process<br> | |
| - | + | response to carbohydrate<br> | |
| - | + | response to hormone<br> | |
| - | + | ||
| - | + | ||
carbohydrate metabolic process<br> | carbohydrate metabolic process<br> | ||
| - | + | protein tetramerization<br> | |
| - | + | regulation of NADP metabolic process<br> | |
| - | + | NADP biosynthetic process<br> | |
| - | + | small molecule metabolic process<br> | |
| + | cellular lipid metabolic process<br> | ||
== In structures == | == In structures == | ||
| - | + | NADP-dependent malic enzyme is found in 3 PDB entries<br> | |
| - | *<b>Eukaryota</b> ( | + | *<b>Eukaryota</b> (3 PDB entries): <br> |
| - | **<b>Homo sapiens</b> ( | + | **<b>Homo sapiens</b> (2 PDB entries): <br> |
| - | ***[[ | + | ***[[3wja]] <div class="pdb-prints 3wja"></div> |
| - | ***Title: The | + | ***Title: The crystal structure of human cytosolic NADP(+)-dependent malic enzyme in apo form |
| - | + | ***2.548 A resolution | |
| - | + | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Homo%20sapiens%22&all_molecule_names:%22NADP-dependent%20malic%20enzyme%22&!chimera:y Search for other PDB entries] | |
| - | + | **<b>Columba livia</b> (1 PDB entries): <br> | |
| - | + | ***[[1gq2]] <div class="pdb-prints 1gq2"></div> | |
| - | + | ***Title: MALIC ENZYME FROM PIGEON LIVER | |
| - | + | ||
| - | *** | + | |
| - | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:% | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **<b> | + | |
| - | ***[[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ***Title: | + | |
***2.5 A resolution | ***2.5 A resolution | ||
| - | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Bombyx%20mori%22&all_molecule_names:%22Lysozyme%22 Search for other PDB entries] | ||
| - | *<b>Viruses</b> (578 PDB entries): <br> | ||
| - | **<b>Enterobacteria phage P22</b> (2 PDB entries): <br> | ||
| - | ***[[2anv]] <div class="pdb-prints 2anv"></div> | ||
| - | ***Title: crystal structure of P22 lysozyme mutant L86M | ||
| - | ***1.04 A resolution | ||
| - | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Enterobacteria%20phage%20P22%22&all_molecule_names:%22Lysozyme%22 Search for other PDB entries] | ||
| - | **<b>Enterobacteria phage P21</b> (2 PDB entries): <br> | ||
| - | ***[[3hdf]] <div class="pdb-prints 3hdf"></div> | ||
| - | ***Title: Crystal structure of truncated endolysin R21 from phage 21 | ||
| - | ***1.7 A resolution | ||
| - | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Enterobacteria%20phage%20P21%22&all_molecule_names:%22Lysozyme%22 Search for other PDB entries] | ||
| - | **<b>Enterobacteria phage T4</b> (561 PDB entries): <br> | ||
| - | ***[[1swy]] <div class="pdb-prints 1swy"></div> | ||
| - | ***Title: Use of a Halide Binding Site to Bypass the 1000-atom Limit to Ab initio Structure Determination | ||
| - | ***1.06 A resolution | ||
| - | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Enterobacteria%20phage%20T4%22&all_molecule_names:%22Lysozyme%22 Search for other PDB entries] | ||
| - | **<b>Escherichia phage T5</b> (1 PDB entries): <br> | ||
| - | ***[[2mxz]] <div class="pdb-prints 2mxz"></div> | ||
| - | ***Title: Bacteriophage T5 l-alanoyl-d-glutamate peptidase comlpex with Zn2+ (Endo T5-ZN2+) | ||
| - | ***N/A A resolution | ||
| - | **<b>Enterobacteria phage lambda</b> (3 PDB entries): <br> | ||
| - | ***[[1am7]] <div class="pdb-prints 1am7"></div> | ||
| - | ***Title: Lysozyme from bacteriophage lambda | ||
| - | ***2.3 A resolution | ||
| - | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Enterobacteria%20phage%20lambda%22&all_molecule_names:%22Lysozyme%22 Search for other PDB entries] | ||
| - | **<b>Streptococcus phage CP-7</b> (1 PDB entries): <br> | ||
| - | ***[[4cvd]] <div class="pdb-prints 4cvd"></div> | ||
| - | ***Title: Crystal structure of the central repeat of cell wall binding module of Cpl7 | ||
| - | ***1.666 A resolution | ||
| - | **<b>Enterobacteria phage P1</b> (2 PDB entries): <br> | ||
| - | ***[[1xju]] <div class="pdb-prints 1xju"></div> | ||
| - | ***Title: Crystal structure of secreted inactive form of P1 phage endolysin Lyz | ||
| - | ***1.07 A resolution | ||
| - | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Enterobacteria%20phage%20P1%22&all_molecule_names:%22Lysozyme%22 Search for other PDB entries] | ||
| - | **<b>Streptococcus phage Cp-1</b> (6 PDB entries): <br> | ||
| - | ***[[2j8g]] <div class="pdb-prints 2j8g"></div> | ||
| - | ***Title: Crystal structure of the modular Cpl-1 endolysin complexed with a peptidoglycan analogue (E94Q mutant in complex with a tetrasaccharide- pentapeptide) | ||
| - | ***1.69 A resolution | ||
| - | ***[http://www.ebi.ac.uk/pdbe/entry/search/index?organism_scientific_name:%22Streptococcus%20phage%20Cp-1%22&all_molecule_names:%22Lysozyme%22 Search for other PDB entries] | ||
| - | *<b>Bacteria</b> (1 PDB entries): <br> | ||
| - | **<b>Enterobacteria phage T4</b> (1 PDB entries): <br> | ||
| - | ***[[2qb0]] <div class="pdb-prints 2qb0"></div> | ||
| - | ***Title: Structure of the 2TEL crystallization module fused to T4 lysozyme with an Ala-Gly-Pro linker. | ||
| - | ***2.56 A resolution | ||
| - | ***Other macromolecules also in this entry: Transcription factor ETV6, . | ||
| - | **<b>Escherichia coli</b> (1 PDB entries): <br> | ||
| - | ***[[2qb0]] <div class="pdb-prints 2qb0"></div> | ||
| - | ***Title: Structure of the 2TEL crystallization module fused to T4 lysozyme with an Ala-Gly-Pro linker. | ||
| - | ***2.56 A resolution | ||
| - | ***Other macromolecules also in this entry: Transcription factor ETV6, . | ||
Revision as of 14:55, 11 September 2015
NADP-dependent malic enzyme
| |||||||||||