Ivan Koutsopatriy estrogen receptor
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
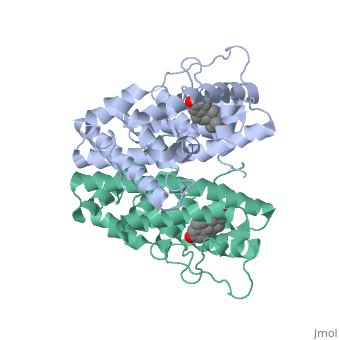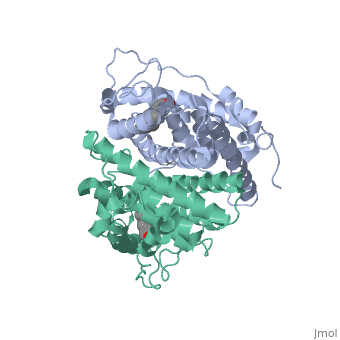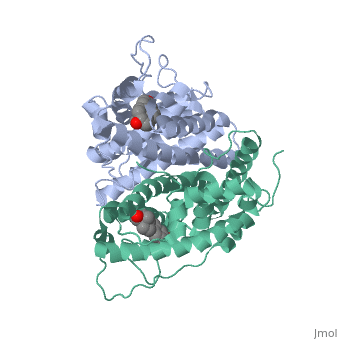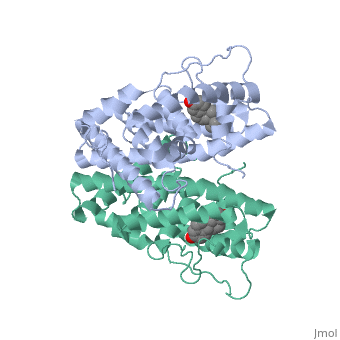
Ferutinine also causes ER to form a tight loop allowing stimulation of normal growth. | Ferutinine also causes ER to form a tight loop allowing stimulation of normal growth. | ||
| - | <scene name='71/714947/Partial_agonist_genistein_er/ | + | <scene name='71/714947/Partial_agonist_genistein_er/3'>Partial_Agonist_genistein_bound_Er</scene> |
The conformation of ER bound to genistein has a loop which is not as tight as those found on ER with a complete agonist ligand. | The conformation of ER bound to genistein has a loop which is not as tight as those found on ER with a complete agonist ligand. | ||
| - | <scene name='71/714947/Antagonist_tamoxifen_bound_er/1'>Antagonist_tamoxifen_bound_er</scene> | ||
| + | <scene name='71/714947/Antagonist_tamoxifen_bound_er/5'>Antagonist_tamoxifen_bound_er</scene> | ||
Tamoxifen is a drug created to bind ER and inhibit the transcription factor activity of ER. Tamoxifen is larger than the normal hormone ER binds (estradiol); for this reason the activation loop is pushed into an inactive conformation. This blocks ER from giving the signal to grow. | Tamoxifen is a drug created to bind ER and inhibit the transcription factor activity of ER. Tamoxifen is larger than the normal hormone ER binds (estradiol); for this reason the activation loop is pushed into an inactive conformation. This blocks ER from giving the signal to grow. | ||
==DNA Protein Interaction and ER Regulation== | ==DNA Protein Interaction and ER Regulation== | ||
| - | <scene name='71/714947/Er_bound_to_dna/ | + | <scene name='71/714947/Er_bound_to_dna/3'>ER_bound_to_dna</scene> |
ER is functional as a ligand-dependent transcription factor. (3) ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. The location of the receptor bound and unbound to ligand varies amongst different cell types. In general, an antagonist ligand will cause partial accumulation in the cytoplasm of a cell. The agonist ligand causes the translocation to the nucleus described above. | ER is functional as a ligand-dependent transcription factor. (3) ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. The location of the receptor bound and unbound to ligand varies amongst different cell types. In general, an antagonist ligand will cause partial accumulation in the cytoplasm of a cell. The agonist ligand causes the translocation to the nucleus described above. | ||
Revision as of 23:28, 23 October 2015
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644