From Proteopedia
(Difference between revisions)
proteopedia linkproteopedia link
|
|
| Line 9: |
Line 9: |
| | | | |
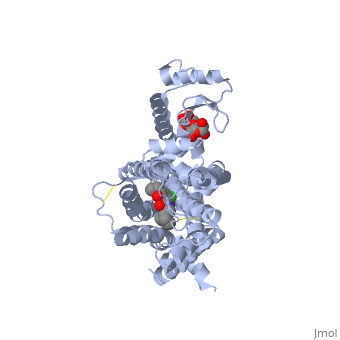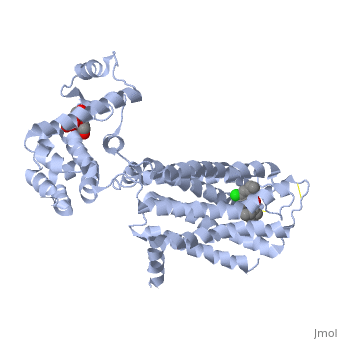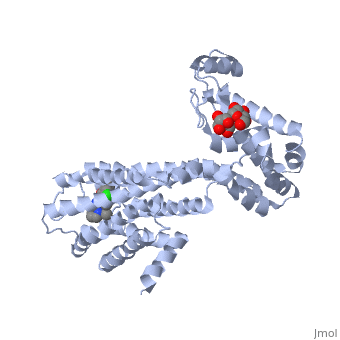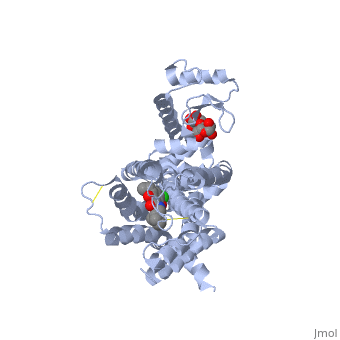
| | Proteins are a long chain of various amino acid sequences. Inside the binding pocket, the residues that are used to bind dopamine to the receptor are Asp-114, Ser-193, Ser-197, Phe-110, Met-117, Cys-118, Phe-164, Phe-189, Val-190, Trp-386, Phe-390, and Hist-394. Dopamine binds to the top of the receptor structure. The pocket formed by these residues is hydrophobic and the residues are consistent among human dopamine receptors (Kalani et al., 2004). In the dopamine receptor, both the N-terminus (amino end of the polypeptide chain) and the C-terminus (carboxyl-group end of the polypeptide chain) are located on the extracellular portions of the cell membrane. Furthermore, all dopamine receptors contain two cysteine amino acids on their extracellular portions whose disulfide bridge helps stabilize this protein (Missale et al., 1998). | | Proteins are a long chain of various amino acid sequences. Inside the binding pocket, the residues that are used to bind dopamine to the receptor are Asp-114, Ser-193, Ser-197, Phe-110, Met-117, Cys-118, Phe-164, Phe-189, Val-190, Trp-386, Phe-390, and Hist-394. Dopamine binds to the top of the receptor structure. The pocket formed by these residues is hydrophobic and the residues are consistent among human dopamine receptors (Kalani et al., 2004). In the dopamine receptor, both the N-terminus (amino end of the polypeptide chain) and the C-terminus (carboxyl-group end of the polypeptide chain) are located on the extracellular portions of the cell membrane. Furthermore, all dopamine receptors contain two cysteine amino acids on their extracellular portions whose disulfide bridge helps stabilize this protein (Missale et al., 1998). |
| | + | |
| | + | == Function == |
| | | | |
| | == Ligands == | | == Ligands == |
Revision as of 22:01, 16 November 2015
Dopamine Receptor
| This is a default text for your page Sandbox1595. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Structure
There are 5 classes of dopamine receptors (D1, D2, D3, D4, D5), all of which have similar structures since they are all part of a G-protein coupled receptor family. This means that the portion of the receptor spanning the inner part of the cell’s membrane is composed of seven membrane-spanning G-protein (guanine nucleotide binding) domains (Beaulieu).
The D1 and D5 dopamine receptors are 80% homologous in their transmembrane domains, whereas the D3 and D4 dopamine receptors are 75 and 53% homologous, respectively, with the D2 receptor (Beaulieu). Meaning, all of the dopamine receptors have similar structures.
Proteins are a long chain of various amino acid sequences. Inside the binding pocket, the residues that are used to bind dopamine to the receptor are Asp-114, Ser-193, Ser-197, Phe-110, Met-117, Cys-118, Phe-164, Phe-189, Val-190, Trp-386, Phe-390, and Hist-394. Dopamine binds to the top of the receptor structure. The pocket formed by these residues is hydrophobic and the residues are consistent among human dopamine receptors (Kalani et al., 2004). In the dopamine receptor, both the N-terminus (amino end of the polypeptide chain) and the C-terminus (carboxyl-group end of the polypeptide chain) are located on the extracellular portions of the cell membrane. Furthermore, all dopamine receptors contain two cysteine amino acids on their extracellular portions whose disulfide bridge helps stabilize this protein (Missale et al., 1998).
Function
Ligands
Disease and Relevance
In the United States, approximately 9.3% of people 12 years old and above struggle from some form of addiction (Treatment Statistics, 2011). Addiction is not only detrimental to the social and behavioral aspects of the person’s life, but it also affects their brain chemistry. Specifically, the use of a wide range of substances like alcohol and illicit drugs interact with dopamine receptors and induce an abnormally high level of Dopamine to flood the brain. Because Dopamine is the primary neurotransmitter in the human reward pathway, the brain begins to associate alcohol or other substance that cause the influx with the huge chemical reward. Initial use is usually caused by the desire to obtain their positive reaction but addiction occurs when the brain no longer functions optimally without the dopamine surge caused by the substance. Because addiction is not only caused by psychological desire but also biological desire it rapidly becomes a detrimental disease to those who suffer.
Malfunction of dopamine receptors also plays a role in a wide array of other neurological problems. There is a delicate balance that must be maintained for the human brain to function at the peak of it’s ability. For example, the psychological disorder schizophrenia is believed to be caused in part by a malfunction in multiple dopamine receptors that result in much higher than normal levels of dopamine, whereas the debilitating disease Parkinson’s is believed to caused in part by the dopamine receptors failing to release a sufficient amount of the neurotransmitter. In the case of schizophrenia it is currently accepted that a wide range of the positive symptoms, including hallucinations and delusions, originate because of an increased level of subcortical dopamine which in turn augments the D2 receptors and leads to even more release in areas of the brain like the nucleus accumbens. Some of the negative effects which include inability to form sentences and lack of outward motivation are hypothesized to be triggered by the reduced activation of D1 receptors (Brisch et al, 201). Parkinson’s, on the other hand, is caused in part by the destruction of dopamine receptors and thus the loss of a critical amount of the neurotransmitter. Dopamine is vital in relaying messages from the brain to the muscular system and disrupting this mechanisms produces tremors and a lack of balance which are common symptoms of the disease (Kim, 2002).
|
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
3. Beaulieu, Jean-Martin, and Gainetdinov, Raul R. "The Physiology, Signaling, and Pharmacology of Dopamine Receptors." Pharmacological Reviews 63.1 (2011): 182-217.
4. Brisch, Ralf, Arthur Saniotis, Rainer Wolf, Hendrik Bielau, Hans-Gert Bernstein, Johann Steiner, Bernhard Bogerts, Katharina Braun, Zbigniew Jankowski, Jaliya Kumaratilake, Maciej Henneberg, and Tomasz Gos. "The Role of Dopamine in Schizophrenia from a Neurobiological and Evolutionary Perspective: Old Fashioned, but Still in Vogue." Frontiers in Psychiatry. Frontiers Media S.A., 19 May 2014. Web. 16 Nov. 2015.
5. "Dopamine." Dopamine. PubChem, 24 Oct. 2015. Web. 27 Oct. 2015.
6. Kalani, M., Vaidehi, N., Hall, S., Trabanino, R., Freddolino, P., Kalani, M., Floriano, W., Kam, V., Goddard, W.(2004). The predicted 3D structure of the human D2 dopamine receptor and the binding site and binding affinities for agonists and antagonists. Proceedings of the National Academy of Sciences, 101(11), 3815-3820. doi:10.1073/pnas.0400100101
7. Kim, Jong-Hoon, and Johnathan Auerbach. "Dopamine Neurons Derived from Embryonic Stem Cells Function in an Animal Model of Parkinson's Disease." Nature.com. Nature Publishing Group, 4 July 2002. Web. 16 Nov. 2015.
8. Missale, C., Nash, S., Robinson, S., Jaber, M., & Caron, M. (1998). Dopamine Receptors: From Structure to Function. Physiological Reviews, 78(1), 189-225. 16 Nov 2015.
9. "Treatment Statistics." DrugFacts:. NIH, Mar. 2011. Web. 16 Nov. 2015.