DNA glycosylase
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
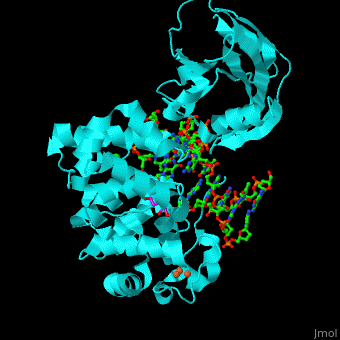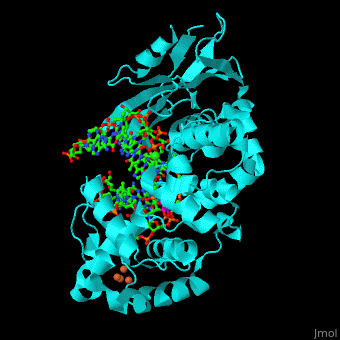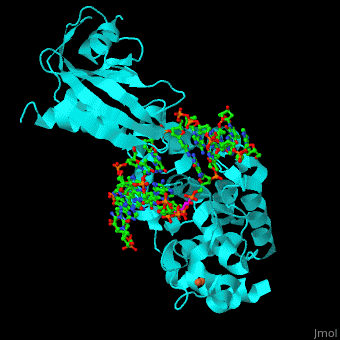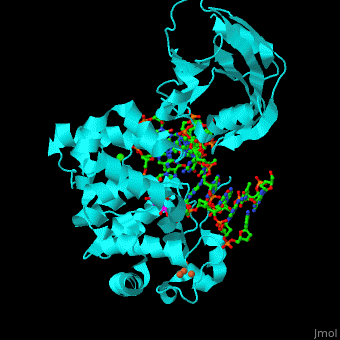
MutY uses base flipping to twist the mispaired adenine out of the DNA helix and into the MutY active site. MutY contains 2 domains: C-terminal domain and catalytic domain containing a helix-hairpin-helix. | MutY uses base flipping to twist the mispaired adenine out of the DNA helix and into the MutY active site. MutY contains 2 domains: C-terminal domain and catalytic domain containing a helix-hairpin-helix. | ||
| - | *<scene name='44/445369/Cv/ | + | *<scene name='44/445369/Cv/4'>Adenine DNA glycosylase containing Fe4S4 cluster with DNA containing an mispaired adenine 8OG and Ca+2</scene>. |
*<scene name='44/445369/Cv/3'>Mispaired adenine 8OG binding site</scene> (PDB code [[1rrs]]).<ref>PMID:14961129</ref> | *<scene name='44/445369/Cv/3'>Mispaired adenine 8OG binding site</scene> (PDB code [[1rrs]]).<ref>PMID:14961129</ref> | ||
</StructureSection> | </StructureSection> | ||
Revision as of 10:13, 6 December 2015
| |||||||||||
3D Structures of DNA glycosylate
Updated on 06-December-2015
References
- ↑ Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004 Feb;14(1):43-9. PMID:15102448 doi:http://dx.doi.org/10.1016/j.sbi.2004.01.003
- ↑ Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004 Feb 12;427(6975):652-6. PMID:14961129 doi:10.1038/nature02306
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky