Gunnar Reiske/Sandbox 102
From Proteopedia
m |
|||
| Line 29: | Line 29: | ||
=== Interactions === | === Interactions === | ||
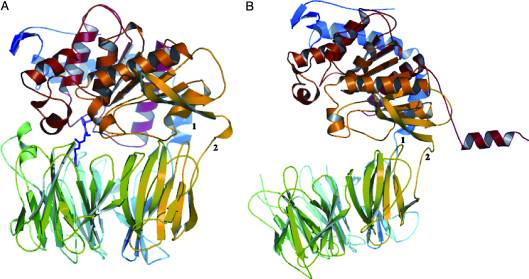
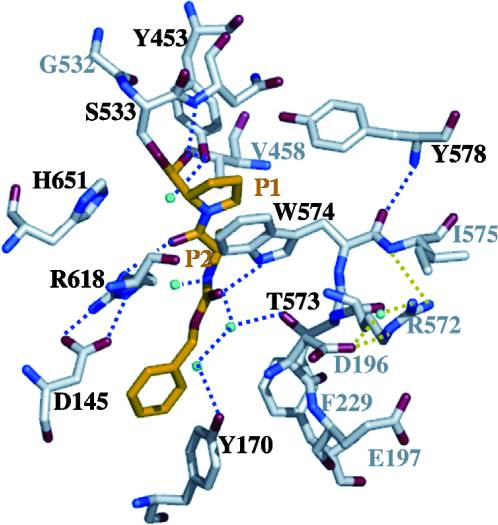
| - | The gluten protein complex binds the HLA-DQ2 complex with hydrogen bonds. Glutamine and lysine hydrogen bond to water, the nitrogen backbone of gliadin, and the asparagine, lysine, tyrosine and serine hydrogens of HLA-DQ2. The proline amino acids are all exposed to the external environment and do not participate in hydrogen binding while also preventing other amino acids from hydrogen binding through steric hindrance. Only proline rich complexes are able to hydrogen bond properly with HLA-DQ2 | + | The gluten protein complex binds the HLA-DQ2 complex with hydrogen bonds. Glutamine and lysine hydrogen bond to water, the nitrogen backbone of gliadin, and the asparagine, lysine, tyrosine and serine hydrogens of HLA-DQ2. The proline amino acids are all exposed to the external environment and do not participate in hydrogen binding while also preventing other amino acids from hydrogen binding through steric hindrance. Only proline rich complexes are able to hydrogen bond properly with HLA-DQ2. All other proteins do not have the proper conformation and organization of proline to effectively bind HLA-DQ2, making the HLA-DQ2-gliadin complex very specific.<ref>Kim, C., Quartsen, H., Bergsen, E., Khosla, C., & Sollid, L.. (n.d.). Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Cross Mark, 101(12), 4175-4179. March 2004 http://www.pnas.org/content/101/12/4175.figures-only</ref> |
Revision as of 01:59, 8 December 2015
How Gluten Protein Structure Stimulates an Immune Response
Introduction
The protein gluten is found in wheat and grains such as rye and barley. Gluten is also involved with inducing an inflammatory response in individuals with celiac disease. Individuals who have the disease cannot digest gluten due to the structure of the protein, which will damage the small intestine. If an individual with celiac disease ingests foods containing gluten, the immune system responds by damaging the villi, which are fingerlike projections lining the small intestine. This immune response reduces the body’s ability to absorb nutrients that pass through the small intestine and into the bloodstream. As a result of the damaged villi, people with celiac disease can become malnourished. Although celiac disease is genetic, the research reviewed for this study placed emphasis on how the protein triggers an immune response in the gastrointestinal tract of affected individuals.
Gluten is a protein complex comprised of gliadin and glutenin. Gliadins, for those with celiac disease, are the principle toxic component of gluten and are composed of proline and glutamine-rich peptide sequences. The peptides enter the circulatory system and come into contact with lymphocytes and T-cells, resulting in the release of cytokines. The cytokines interact with the villi of the small intestine and damage them, disabling the body from nutrient absorption. The symptoms can include abdominal pain, weight loss, fatigue, and many other symptoms associated with malnutrition. As of now, the only treatment for celiac disease is the total exclusion of gluten from the person’s diet.[1][2][3]
| |||||||||||
References
- ↑ Celiac Disease: MedlinePlus. Retrieved October 27, 2015, from https://www.nlm.nih.gov/medlineplus/celiacdisease.html
- ↑ Celiac Disease. Retrieved October 27, 2015, from http://www.niddk.nih.gov/health-information/health-topics/digestive-diseases/celiac-disease/Pages/facts.aspx
- ↑ Go to Science. Retrieved October 27, 2015, from http://www.sciencemag.org/content/297/5590/2275.full
- ↑ 4.0 4.1 Maiuri, L., Ciacci, C., Ricciardelli, I., Vacca, L., Raia, V., Auricchio, S., . . . Londei, M. (2003). Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet, 362(9377), 30-37. doi:10.1016/S0140-6736(03)13803-2
- ↑ Mellins, E., & Stern, L. (n.d.). HLA-DM and HLA-DO, key regulators of MHC-ll processing and presentation. Current Opinion in Immunology, 26, 115-122. February 2014. http://www.sciencedirect.com/science/article/pii/S095279151300215X
- ↑ Kim, C., Quartsen, H., Bergsen, E., Khosla, C., & Sollid, L.. (n.d.). Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Cross Mark, 101(12), 4175-4179. March 2004 http://www.pnas.org/content/101/12/4175.figures-only
- ↑ Henderson, K. N., Tye-Din, J. A., Reid, H. H., Chen, Z., Borg, N. A., Beissbarth, T., . . . Anderson, R. P. (2007). A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity,27(1), 23-34. doi:10.1016/j.immuni.2007.05.015
- ↑ 8.0 8.1 8.2 8.3 8.4 Shan, L., I. I. Mathews, and C. Khosla. "Structural and Mechanistic Analysis of Two Prolyl nEndopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity." Proceedings of the National Academy of Sciences 102.10 (2005): 3599-604. Web.
- ↑ Matysiak-Budnik, T., Candalh, C., Cellier, C., Dugave, C., Namane, A., Vidal-Martinez, T., . . . Heyman, M. (2005). Limited efficiency of prolyl-endopeptidase in the detoxification of gliadin peptides in celiac disease. Gastroenterology,129(3), 786-796. doi:10.1053/j.gastro.2005.06.016
Proteopedia Page Contributors and Editors (what is this?)
Ben Horansky, Devin Joseph, Premal Patel, Gunnar Reiske, Katlin Cannon