We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Clp Protease
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | |||
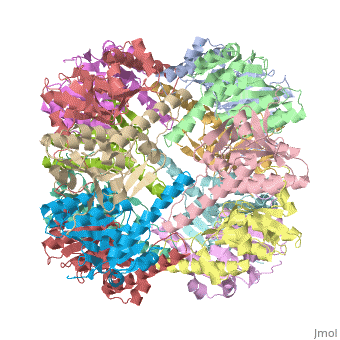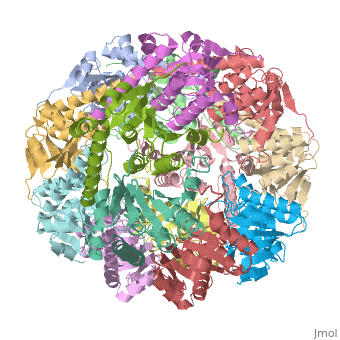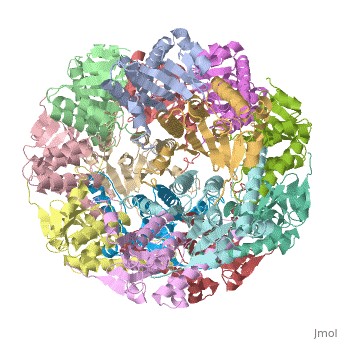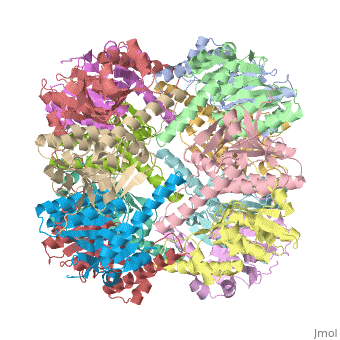
<StructureSection load='1tyf' size='350' side='right' caption='E. coli Clp protease (PDB entry [[1tyf]])' scene=''> | <StructureSection load='1tyf' size='350' side='right' caption='E. coli Clp protease (PDB entry [[1tyf]])' scene=''> | ||
| - | '''Clp protease''' (CLP) is a serine peptidase which hydrolyzes proteins in the presence of ATP and Mg+2 ion. CLP is found in mitochondria. CLP participates in degradation of misfolded proteins | + | == Function == |
| + | |||
| + | '''Clp protease''' (CLP) is a serine peptidase which hydrolyzes proteins in the presence of ATP and Mg+2 ion. CLP is found in mitochondria. CLP participates in degradation of misfolded proteins. For more details see <br /> | ||
* [[Clp protease]] | * [[Clp protease]] | ||
* [[Molecular Playground/ClpP]]. | * [[Molecular Playground/ClpP]]. | ||
| + | |||
| + | == Structural highlights == | ||
| + | |||
| + | CLP is a heterodimer containing an ATP-binding regulatory subunit A and catalytic subunit P. | ||
</StructureSection> | </StructureSection> | ||
==3D structures of Clp protease== | ==3D structures of Clp protease== | ||
Revision as of 11:30, 8 December 2015
| |||||||||||
3D structures of Clp protease
Updated on 08-December-2015