Dioxygenase
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
* '''Naphthalene 1,2-dioxygenase''' (NDO) catalyzes the conversion of naphthalene and O2 to dihydronaphthalene-diol.<br /> | * '''Naphthalene 1,2-dioxygenase''' (NDO) catalyzes the conversion of naphthalene and O2 to dihydronaphthalene-diol.<br /> | ||
* '''Glyoxalase/bleomycin resistant protein/dioxygenase''' (GBD) catalyzes the cleavage of S-lactoyl-glutathione to methylglyoxal and glutathione.<br /> | * '''Glyoxalase/bleomycin resistant protein/dioxygenase''' (GBD) catalyzes the cleavage of S-lactoyl-glutathione to methylglyoxal and glutathione.<br /> | ||
| - | * '''α-ketoglutarate-dependent dioxygenase homolog 5''' (Alkbh5) is an RNA demethylase which reverses the N(6)-methyladenosine modification of RNA. | + | * '''α-ketoglutarate-dependent dioxygenase homolog 5''' (Alkbh5) is an RNA demethylase which reverses the N(6)-methyladenosine modification of RNA.<br /> |
| + | * '''Ten-eleven translocation methylcytosine dioxygenase''' (TET) catalyzes conversion of 5-methylcytosine to 5-hydroxymethylcytosine<ref>PMID:25862091</ref>. For details see [[TET Enzymes]]. | ||
=== A Hyperactive Cobalt-Substituted Extradiol-Cleaving Catechol Dioxygenase <ref>DOI 10.1007/s00775-010-0732-0</ref>=== | === A Hyperactive Cobalt-Substituted Extradiol-Cleaving Catechol Dioxygenase <ref>DOI 10.1007/s00775-010-0732-0</ref>=== | ||
| Line 140: | Line 141: | ||
**[[1gp5]], [[1gp6]] – AtLACD + dihydroquercetin – ''Arabidopsis thaliana'' <br /> | **[[1gp5]], [[1gp6]] – AtLACD + dihydroquercetin – ''Arabidopsis thaliana'' <br /> | ||
**[[2brt]] – AtLACD + naringenin <br /> | **[[2brt]] – AtLACD + naringenin <br /> | ||
| + | |||
| + | *Ten-eleven translocation methylcytosine dioxygenase (TET) | ||
| + | |||
| + | **[[5deu]] - hTET2 + DNA + deoxyhydroxymethyl cytidine dihydrogen phosphate<br /> | ||
| + | **[[5d9y]] - hTET2 + DNA + formyl deoxy CMP<br /> | ||
| + | **[[4nm6]] - hTET2 + DNA + methyl deoxy CMP<br /> | ||
| + | **[[4z3c]] - hTET3 + DNA<br /> | ||
| + | **[[5exh]] - mTET3 CXXC domain + DNA + carboxy deoxy CMP<br /> | ||
*'''Dioxygenase extradiol family''' | *'''Dioxygenase extradiol family''' | ||
Revision as of 16:21, 13 January 2017
| |||||||||||
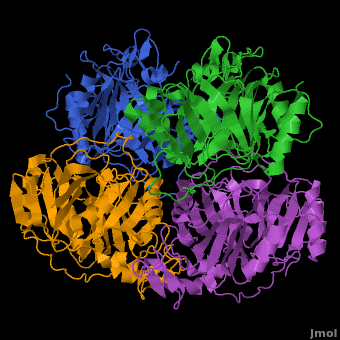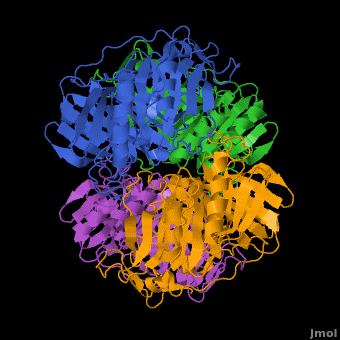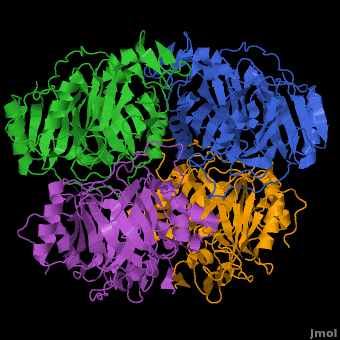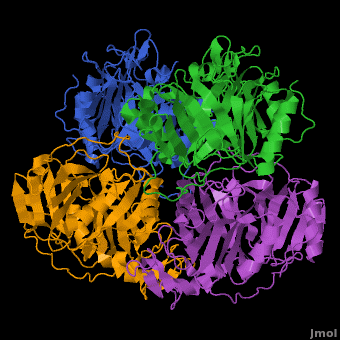
3D structures of Protocatechuate 3,4-dioxygenase
Updated on 13-January-2017
- ↑ Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, Tian Q, Aifantis I, Wei L, Dong C. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015 Apr 21;42(4):613-26. doi: 10.1016/j.immuni.2015.03.005. Epub 2015 , Apr 7. PMID:25862091 doi:http://dx.doi.org/10.1016/j.immuni.2015.03.005
- ↑ Fielding AJ, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L Jr. A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase. J Biol Inorg Chem. 2010 Dec 14. PMID:21153851 doi:10.1007/s00775-010-0732-0
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky