D-xylose isomerase
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Structural highlights == | == Structural highlights == | ||
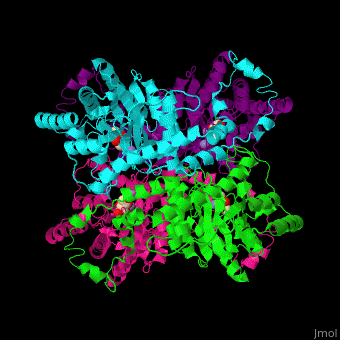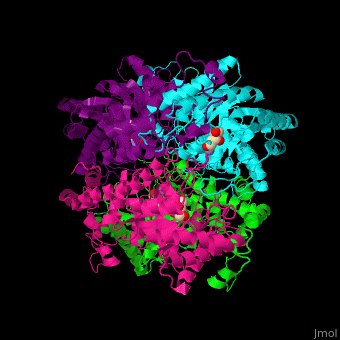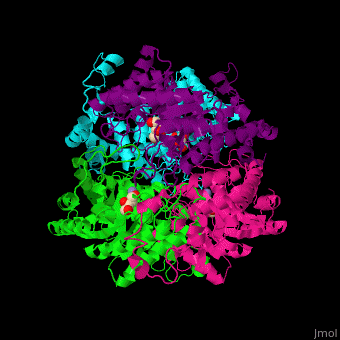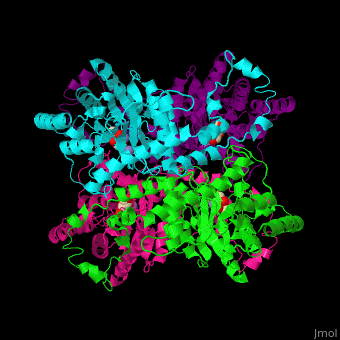
| - | <scene name='52/525128/Cv/2'>The biological assembly of D-xylose isomerase</scene> from ''Arthrobacter sp'' is homotetramer. DXI active site contains <scene name='52/525128/Cv/4'>2 Mn+2 ions</scene> and the <scene name='52/525128/Cv/6'>ligand binding pocket is composed of aromatic and acidic residues</scene>.<ref>PMID:2319597</ref> | + | <scene name='52/525128/Cv/2'>The biological assembly of D-xylose isomerase</scene> from ''Arthrobacter sp'' is homotetramer. DXI active site contains <scene name='52/525128/Cv/4'>2 Mn+2 ions</scene> and the <scene name='52/525128/Cv/6'>ligand binding pocket is composed of aromatic and acidic residues</scene> (water molecules shown as red spheres).<ref>PMID:2319597</ref> |
</StructureSection> | </StructureSection> | ||
==3D structures of D-xylose isomerase== | ==3D structures of D-xylose isomerase== | ||
Revision as of 11:50, 11 January 2016
| |||||||||||
3D structures of D-xylose isomerase
Updated on 11-January-2016
References
- ↑ MARSHALL RO, KOOI ER. Enzymatic conversion of D-glucose to D-fructose. Science. 1957 Apr 5;125(3249):648-9. PMID:13421660
- ↑ Collyer CA, Henrick K, Blow DM. Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift. J Mol Biol. 1990 Mar 5;212(1):211-35. PMID:2319597