Thrombin
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
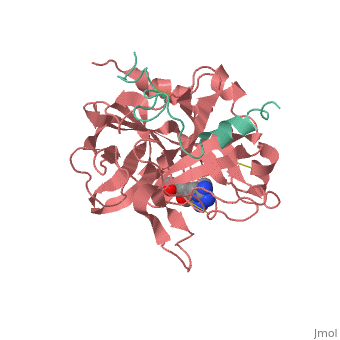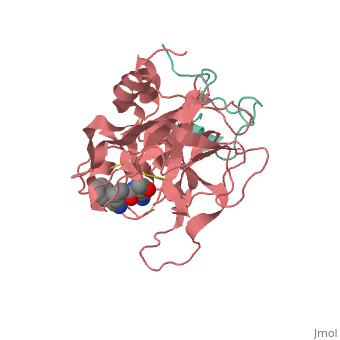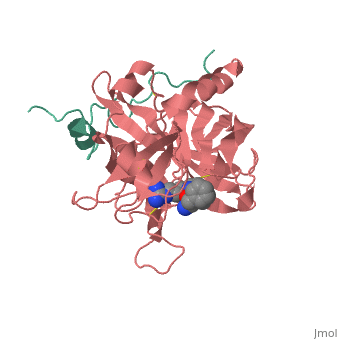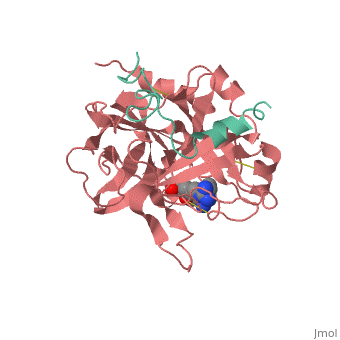
<StructureSection load='1ppb' size='350' side='right' scene='' caption='Human thrombin large (red) and small (aqua) subunits complex with prolinamide derivative (PDB code [[1ppb]])'> | <StructureSection load='1ppb' size='350' side='right' scene='' caption='Human thrombin large (red) and small (aqua) subunits complex with prolinamide derivative (PDB code [[1ppb]])'> | ||
| + | ==Introduction== | ||
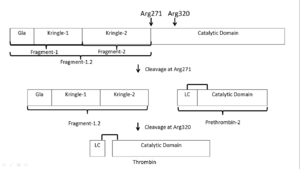
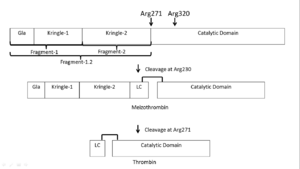
'''Thrombin''' (Thr) is a serine protease. '''Prothrombin''' (PThr) is cleaved to form Thr in the coagulation cascade. The first step of the cleavage is at residue R320 and produces '''meizothrombin''' (MThr). Thr catalyzes the conversion of fibrinogen to the insoluble fibrin. Thr is composed of heavy chain (HC) and light chain (LC). Prethrombin-1 lacks 155 N-terminal residues of PThr and is composed of a single polypeptide chain. '''Prethrombin-2''' is the product of proteolysis of '''prethrombin-1''' by trypsin or by active factor X. P-PACK Thr is a chemicaly modified Thr with inactivated catalytic site and active anion binding site. Hirudin is the most potent natural inhibitor of Thr ([[Sean Swale/Human Thrombin Inhibitor]]). For some more details see [[Serine Proteases]]. Prothrombin cleavage results in the creation of thrombin, a coagulative agent in plasma and is connected to fibrinolysis and platelet activation. During this process several peptides involved in the conversion are released into the plasma, and the remaining protein splits into two portions(http://www.uniprot.org/citations/3759958). It has been shown that prothrombin has a statistically significant connection to the occurrence of ischemic stroke with the presence of the G20210A mutation, though the cause was not isolated to prothrombin alone (http://www.uniprot.org/citations/15534175) (these links added by Connor Gramazio). Some additional details in<br /> | '''Thrombin''' (Thr) is a serine protease. '''Prothrombin''' (PThr) is cleaved to form Thr in the coagulation cascade. The first step of the cleavage is at residue R320 and produces '''meizothrombin''' (MThr). Thr catalyzes the conversion of fibrinogen to the insoluble fibrin. Thr is composed of heavy chain (HC) and light chain (LC). Prethrombin-1 lacks 155 N-terminal residues of PThr and is composed of a single polypeptide chain. '''Prethrombin-2''' is the product of proteolysis of '''prethrombin-1''' by trypsin or by active factor X. P-PACK Thr is a chemicaly modified Thr with inactivated catalytic site and active anion binding site. Hirudin is the most potent natural inhibitor of Thr ([[Sean Swale/Human Thrombin Inhibitor]]). For some more details see [[Serine Proteases]]. Prothrombin cleavage results in the creation of thrombin, a coagulative agent in plasma and is connected to fibrinolysis and platelet activation. During this process several peptides involved in the conversion are released into the plasma, and the remaining protein splits into two portions(http://www.uniprot.org/citations/3759958). It has been shown that prothrombin has a statistically significant connection to the occurrence of ischemic stroke with the presence of the G20210A mutation, though the cause was not isolated to prothrombin alone (http://www.uniprot.org/citations/15534175) (these links added by Connor Gramazio). Some additional details in<br /> | ||
* [[Ann Taylor 115]]<br /> | * [[Ann Taylor 115]]<br /> | ||
| Line 6: | Line 7: | ||
* [[Thrombin light chain]]<br /> | * [[Thrombin light chain]]<br /> | ||
| - | + | <scene name='58/583418/Thombin_main_secondary/2'>Thrombin</scene> catalyzes the penultimate step in blood coagulation. It is activated from its [http://en.wikipedia.org/wiki/Zymogen zymogen], prothrombin, at the site of tissue injury by [[Factor_Xa | Factor Xa (FXa)]] and its cofactor [http://en.wikipedia.org/wiki/Factor_V FVa] in the presence of phospholipid membrane and calcium. Thrombin is then able to catalyze the cleavage of [[Fibrinogen | fibrinogen]] to insoluable fibrin which spontaneously polymerizes to form a stable clot.<ref name="zero">PMID: 7023326</ref><ref name="one">PMID: 11001069</ref> Thrombin also acts as a procoagulant by: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <scene name='58/583418/Thombin_main_secondary/2'>Thrombin</scene> | + | |
* Activating platelets through their [http://en.wikipedia.org/wiki/Protease-activated_receptor protease activated receptors (PARs)]<ref name="one"/> | * Activating platelets through their [http://en.wikipedia.org/wiki/Protease-activated_receptor protease activated receptors (PARs)]<ref name="one"/> | ||
| Line 32: | Line 25: | ||
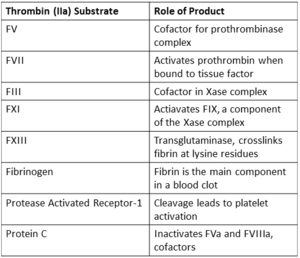
By balancing substrate specificity, activity, and inhibition thrombin plays a central role in the blood coagulation cascade. <ref name="three"/> | By balancing substrate specificity, activity, and inhibition thrombin plays a central role in the blood coagulation cascade. <ref name="three"/> | ||
[[Image:Substrates.png|300px|center|thumb| Coagulation related substrates of thrombin, excluding serpin inhibitors.]] | [[Image:Substrates.png|300px|center|thumb| Coagulation related substrates of thrombin, excluding serpin inhibitors.]] | ||
| + | ==Structure== | ||
| + | |||
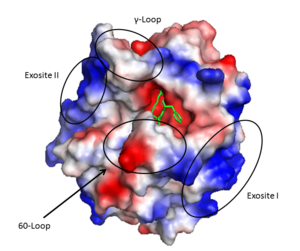
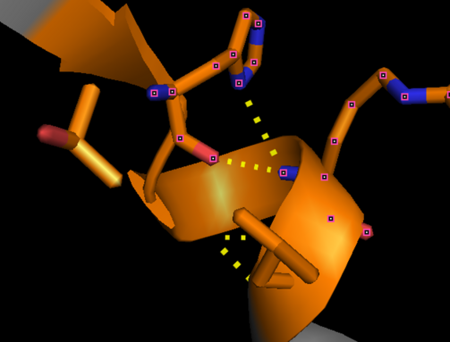
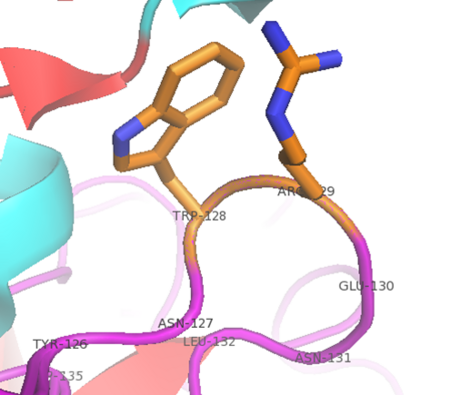
| + | '''Thrombin''' is a "trypsin-like" serine protease. Its structure (PDB code [[1ppb]]) is shown here with a peptide chloroketone inhibitor (PPACK). The thrombin A chain (cleaved N terminal fragement) is shown in cyan and the B chain is shown in red. The <scene name='Serine_Protease/Active_site/2'>Active site</scene> is made up of a catalytic triad of Ser195, His57 and Asp102, backed up by Ser214. The peptide chloroketone inhibitor (PPACK) is shown in purple. A closeup shows the <scene name='Serine_Protease/Activation_site/2'>activation site</scene> at which the sidechain of Asp194 makes a salt link with the N-terminus at residue 16, newly formed when the A chain is cleaved in the zymogen-to-enzyme activation process. The specificity pocket is on one side of the throat of the domain 2 beta barrel, and the activation site is close next to it. | ||
| + | |||
| + | The B chain consists of <scene name='Serine_Protease/Domains/1'>two domains</scene>. As is true for all of the "trypsin-like" serine proteases, each of the two thrombin domains consists mainly of a 6-stranded, antiparallel beta barrel. The specificity pocket (here filled with the Lys sidechain of the PPACK inhibitor) is in one side of the throat of the domain 2beta barrel, and the activation site is close next to it. | ||
| + | |||
| + | Active site residues Ser195, Asp102, and His57 are viewed in ball and stick form.' scene='58/583418/Thombin_main_secondary/2'> | ||
| + | |||
Revision as of 07:47, 25 January 2016
| |||||||||||
3D Structures of thrombin
Updated on 25-January-2016
With participation by Cody Couperus
References
- ↑ Fenton JW 2nd. Thrombin specificity. Ann N Y Acad Sci. 1981;370:468-95. PMID:7023326
- ↑ 2.0 2.1 Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000 Sep 14;407(6801):258-64. PMID:11001069 doi:http://dx.doi.org/10.1038/35025229
- ↑ Crawley JT, Lam JK, Rance JB, Mollica LR, O'Donnell JS, Lane DA. Proteolytic inactivation of ADAMTS13 by thrombin and plasmin. Blood. 2005 Feb 1;105(3):1085-93. Epub 2004 Sep 23. PMID:15388580 doi:http://dx.doi.org/10.1182/blood-2004-03-1101
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Lane DA, Philippou H, Huntington JA. Directing thrombin. Blood. 2005 Oct 15;106(8):2605-12. Epub 2005 Jun 30. PMID:15994286 doi:http://dx.doi.org/10.1182/blood-2005-04-1710
- ↑ Takagi T, Doolittle RF. Amino acid sequence studies on factor XIII and the peptide released during its activation by thrombin. Biochemistry. 1974 Feb 12;13(4):750-6. PMID:4811064
- ↑ Miljic P, Heylen E, Willemse J, Djordjevic V, Radojkovic D, Colovic M, Elezovic I, Hendriks D. Thrombin activatable fibrinolysis inhibitor (TAFI): a molecular link between coagulation and fibrinolysis. Srp Arh Celok Lek. 2010 Jan;138 Suppl 1:74-8. PMID:20229688
- ↑ 7.0 7.1 7.2 7.3 Huntington JA. Natural inhibitors of thrombin. Thromb Haemost. 2014 Apr 1;111(4):583-9. doi: 10.1160/TH13-10-0811. Epub 2014 Jan, 30. PMID:24477356 doi:http://dx.doi.org/10.1160/TH13-10-0811
- ↑ 8.0 8.1 8.2 8.3 8.4 Huntington JA. Thrombin inhibition by the serpins. J Thromb Haemost. 2013 Jun;11 Suppl 1:254-64. doi: 10.1111/jth.12252. PMID:23809129 doi:http://dx.doi.org/10.1111/jth.12252
- ↑ Esmon CT. The regulation of natural anticoagulant pathways. Science. 1987 Mar 13;235(4794):1348-52. PMID:3029867
- ↑ Kalafatis M, Rand MD, Mann KG. The mechanism of inactivation of human factor V and human factor Va by activated protein C. J Biol Chem. 1994 Dec 16;269(50):31869-80. PMID:7989361
- ↑ 11.0 11.1 Lu D, Kalafatis M, Mann KG, Long GL. Comparison of activated protein C/protein S-mediated inactivation of human factor VIII and factor V. Blood. 1996 Jun 1;87(11):4708-17. PMID:8639840
- ↑ Duga S, Asselta R, Tenchini ML. Coagulation factor V. Int J Biochem Cell Biol. 2004 Aug;36(8):1393-9. PMID:15147718 doi:http://dx.doi.org/10.1016/j.biocel.2003.08.002
- ↑ Saenko EL, Shima M, Sarafanov AG. Role of activation of the coagulation factor VIII in interaction with vWf, phospholipid, and functioning within the factor Xase complex. Trends Cardiovasc Med. 1999 Oct;9(7):185-92. PMID:10881749
- ↑ Camire, R. M. (2010). Platelet factor V to the rescue. Blood, 115(4), 753-754. DOI: 10.1182/blood-2009-11-252619
- ↑ Berkner KL. Vitamin K-dependent carboxylation. Vitam Horm. 2008;78:131-56. doi: 10.1016/S0083-6729(07)00007-6. PMID:18374193 doi:http://dx.doi.org/10.1016/S0083-6729(07)00007-6
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 16.6 16.7 Lechtenberg BC, Freund SM, Huntington JA. An ensemble view of thrombin allostery. Biol Chem. 2012 Sep;393(9):889-98. doi: 10.1515/hsz-2012-0178. PMID:22944689 doi:http://dx.doi.org/10.1515/hsz-2012-0178
- ↑ Tijburg PN, van Heerde WL, Leenhouts HM, Hessing M, Bouma BN, de Groot PG. Formation of meizothrombin as intermediate in factor Xa-catalyzed prothrombin activation on endothelial cells. The influence of thrombin on the reaction mechanism. J Biol Chem. 1991 Feb 25;266(6):4017-22. PMID:1995649
- ↑ Bobofchak KM, Pineda AO, Mathews FS, Di Cera E. Energetic and structural consequences of perturbing Gly-193 in the oxyanion hole of serine proteases. J Biol Chem. 2005 Jul 8;280(27):25644-50. Epub 2005 May 12. PMID:15890651 doi:http://dx.doi.org/10.1074/jbc.M503499200
- ↑ 19.0 19.1 19.2 19.3 Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989 Nov;8(11):3467-75. PMID:2583108
- ↑ Page MJ, Di Cera E. Evolution of peptidase diversity. J Biol Chem. 2008 Oct 31;283(44):30010-4. doi: 10.1074/jbc.M804650200. Epub 2008 , Sep 3. PMID:18768474 doi:http://dx.doi.org/10.1074/jbc.M804650200
- ↑ Schechter I, Berger A. On the size of the active site in proteases. I. Papain. 1967. Biochem Biophys Res Commun. 2012 Aug 31;425(3):497-502. doi:, 10.1016/j.bbrc.2012.08.015. PMID:22925665 doi:http://dx.doi.org/10.1016/j.bbrc.2012.08.015
- ↑ Huntington JA. Molecular recognition mechanisms of thrombin. J Thromb Haemost. 2005 Aug;3(8):1861-72. PMID:16102053 doi:http://dx.doi.org/10.1111/j.1538-7836.2005.01363.x
- ↑ Zhang E, Tulinsky A. The molecular environment of the Na+ binding site of thrombin. Biophys Chem. 1997 Jan 31;63(2-3):185-200. PMID:9108691
- ↑ Li W, Johnson DJ, Esmon CT, Huntington JA. Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat Struct Mol Biol. 2004 Sep;11(9):857-62. Epub 2004 Aug 15. PMID:15311269 doi:10.1038/nsmb811
- ↑ Spronk HM, Borissoff JI, ten Cate H. New insights into modulation of thrombin formation. Curr Atheroscler Rep. 2013 Nov;15(11):363. doi: 10.1007/s11883-013-0363-3. PMID:24026641 doi:http://dx.doi.org/10.1007/s11883-013-0363-3
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Cody Couperus, Joel L. Sussman