We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1137
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
=== Cellular effects === | === Cellular effects === | ||
---- | ---- | ||
| - | [[autophagy]] | ||
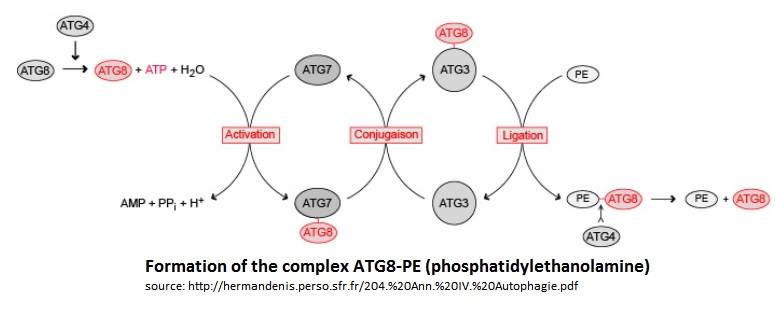
The autophagy related proteins (Atg) are proteins which purpose is to help the parasite to survive and develop. Atg8 and Atg3 are both involved in the [https://en.wikipedia.org/wiki/Autophagy autophagy] process, by creating an autophagosome and then making it grow. The autophagosome can be compared to a vacuole and absorbs the components of the cytoplasm (in a non specific way). When the growing phase (sporozoite) of the cell is over,in the case of ''Plasmodium falciparum'', the autophagy is launched by the cell in order to become erythrocytic-infective. This is the time when the cell starts its viral activity. Lysosomes then merge with the autophagosme to form the autolysosome. Once this process is done, the autolysosome can complete its purpose : recycle the its content. Infact, all the molecules inside are degradated and their components (like aminoacids) are reused in order to synthetise viral proteins. | The autophagy related proteins (Atg) are proteins which purpose is to help the parasite to survive and develop. Atg8 and Atg3 are both involved in the [https://en.wikipedia.org/wiki/Autophagy autophagy] process, by creating an autophagosome and then making it grow. The autophagosome can be compared to a vacuole and absorbs the components of the cytoplasm (in a non specific way). When the growing phase (sporozoite) of the cell is over,in the case of ''Plasmodium falciparum'', the autophagy is launched by the cell in order to become erythrocytic-infective. This is the time when the cell starts its viral activity. Lysosomes then merge with the autophagosme to form the autolysosome. Once this process is done, the autolysosome can complete its purpose : recycle the its content. Infact, all the molecules inside are degradated and their components (like aminoacids) are reused in order to synthetise viral proteins. | ||
=== Molecular effects === | === Molecular effects === | ||
| Line 22: | Line 21: | ||
== Structural highlights == | == Structural highlights == | ||
===Atg3 protein=== | ===Atg3 protein=== | ||
| + | Atg3 is a protein composed of 314 aminoacids with an alpha/beta-fold with a core region topologically similar to E2 enzymes. The core region has two regions: | ||
| + | → the first region has 80 residues and has a random coil structure in solution, this region is responsible for the Atg7 interaction which is an E1-like enzyme. | ||
| + | → The second is an alpha-helical structure which protrudes from the core region. This alpha-helical structure is responsible for binding Atg8. | ||
| + | Moreover some researches indicates that the catalytic cysteine of Atg3 is a possible binding site for a phosphate of phosphatidylethanolamine. | ||
===Atg8 protein=== | ===Atg8 protein=== | ||
| + | Atg8 is a protein of 117 aminoacids with a molecular wieght of 13,6kDa. This molecule is composed of 5- stranded β-sheet. Those β-sheet are enclosed by two α-helices on each sides. This conformation leaves accessible a conserved GABARAP domain, this protein has been originally identified as a binding partner of a GABAA receptor subunit. [https://en.wikipedia.org/wiki/GABARAP GABARAP]. Even if the sequences between Atg8 and ubiquitin are not similars, the crystal structure reveals a conserved ubiquitine-like fold. Atg8 belongs to the ATG family but it differs from the other members of the family because the α2 helix-terminating proline 26 was substituted by a lysine. | ||
===Atg8/Atg3 Complex=== | ===Atg8/Atg3 Complex=== | ||
Revision as of 18:33, 29 January 2016
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Plasmodium falciparum Atg8 in complex with Plasmodium falciparum Atg3 peptide
| |||||||||||
References
http://hermandenis.perso.sfr.fr/204.%20Ann.%20IV.%20Autophagie.pdf