We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1136
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
Sucrose synthase catalyses the cleavage reaction at pH around 7 and the synthesis reaction at pH above 9. Cytoplasmic pH is around 6,5-7, it suggests that SUS works in vivo only to cleave sucrose, which explain why SUS1 is mostly in sink organs. Therefore, it is a linker between all the pathways that use or synthesize sucrose. SUS is involved in biomass production, nitrogen fixation, fruit and seed maturation and can response to stress (cold, anaerobic conditions, drought or gene induction). | Sucrose synthase catalyses the cleavage reaction at pH around 7 and the synthesis reaction at pH above 9. Cytoplasmic pH is around 6,5-7, it suggests that SUS works in vivo only to cleave sucrose, which explain why SUS1 is mostly in sink organs. Therefore, it is a linker between all the pathways that use or synthesize sucrose. SUS is involved in biomass production, nitrogen fixation, fruit and seed maturation and can response to stress (cold, anaerobic conditions, drought or gene induction). | ||
However, the binding to target mechanism is unknown at the molecular level. | However, the binding to target mechanism is unknown at the molecular level. | ||
| - | In a mutant SUS1, we observe a decrease in amidon and lipids concentration, a misbalance between hexose and sucrose, and a perturbation of the organic | + | In a mutant SUS1, we observe a decrease in amidon and lipids concentration, a misbalance between hexose and sucrose, and a perturbation of the organic acids synthesis (citrate and malate), during seed maturation |
<ref>11 Juan Gabriel Angeles N_U ~Nez. Study of sucrose synthase in arabdopsis seed : lacalization, | <ref>11 Juan Gabriel Angeles N_U ~Nez. Study of sucrose synthase in arabdopsis seed : lacalization, | ||
regulation and function. Sciences of the U.</ref>. | regulation and function. Sciences of the U.</ref>. | ||
| Line 43: | Line 43: | ||
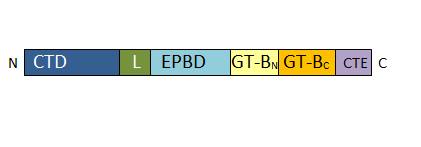
[[Image:Monomer structure.jpg]] | [[Image:Monomer structure.jpg]] | ||
| + | ''Primary structure of Atsus1 monomer. CTD : for Cellular Targeting Domain (residues 0-127); L for Linker (residues 128-156); EPBD: for ENOD40 Peptide-Binding Domain (residues 157-276); GT-B glycosyltransferase : Gt-BN (residues 277-526) and Gt-BC (residues 527-754); CTE for C-Terminal Extension (residues 776-808)'' | ||
The SUS1 tetramer is flat, with two types of subunit interfaces, the A:B and A:D interfaces. The A:D interface is an interaction between the C-terminal extension and the linker, whereas the A:B interface is created by the interaction of adjacent EPBD domains. | The SUS1 tetramer is flat, with two types of subunit interfaces, the A:B and A:D interfaces. The A:D interface is an interaction between the C-terminal extension and the linker, whereas the A:B interface is created by the interaction of adjacent EPBD domains. | ||
| Line 51: | Line 52: | ||
== Regulation == | == Regulation == | ||
| - | ENOD40-A is a small hormon-like peptide able to specifically thiolates the Cys-266 of AtSUS1. It also inhibit the phosphorylation of Ser-167. In Arabidopsis, SUS1 gene is probably regulated by both by sucrose and D-Glucose | + | Sucrose synthase regulation is not well known, but here are some of the supposed mechanisms. |
| + | • ENOD40-A is a small hormon-like peptide able to specifically thiolates the Cys-266 of AtSUS1. It also inhibit the phosphorylation of Ser-167. | ||
| + | • In Arabidopsis, SUS1 gene is probably regulated by both by sucrose and D-Glucose. | ||
| + | • Conformational changes of the EPBD may change the active site activity through distortions of an α helix. | ||
| + | • The CTD can possibly interact with actin and other cellular target, which can regulate sucrose synthase activity. | ||
| + | • In other organisms, it has been shown that the sucrose synthase is active into its dimer form, it is supposed to be the same phenomenon in ''Arabidopsis thaliana'', though it has not been prouved. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 14:22, 30 January 2016
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Structure and Functional aspects of Sucrose Synthase from Arabidopsis thaliana
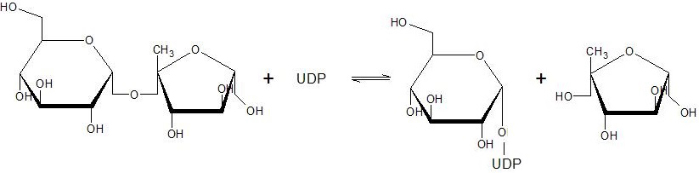
Sucrose Synthase 1 (EC:2.4.1.13), also known as the Sucrose-UDP glucolsyltransferase 1, is a reversible enzyme allowing the synthesis or the degradation of Sucrose in Arabidopsis thaliana. It is a 360 kDa tetramer and belongs to the Glycosyltransferase subfamily 4 (GT4).
| |||||||||||
References
• UniProt entry: P49040
• Brenda entry : 2.4.1.13
- ↑ Salerno GL, Curatti L Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci. 2003 Feb
- ↑ Baroja-Fernández, E., Muñoz, F.J., Saikusa, T., Rodríguez-López, M., Akazawa, T. and Pozueta-Romero, J. Sucrose synthase catalyzes the de novo production of ADPglucose linked to starch biosynthesis in heterotrophic tissues of plants. Plant Cell Physiol.
- ↑ 11 Juan Gabriel Angeles N_U ~Nez. Study of sucrose synthase in arabdopsis seed : lacalization, regulation and function. Sciences of the U.
- ↑ Sucrose synthase oligomerization and F-actin association are regulated by sucrose concentration and phosphorylation. Duncan KA, Huber SC. Plant Cell Physiol. 2007 Nov; 48(11):1612-23.
- ↑ Salerno GL, Curatti L. Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci. 2003 Feb
- ↑ Determination of structural requirements and probable regulatory effectors for membrane association of maize sucrose synthase 1. Hardin SC, Duncan KA, Huber SC. Plant Physiol. 2006
- ↑ Phosphorylation of sucrose synthase at serine 170: occurrence and possible role as a signal for proteolysis. Hardin SC, Tang GQ, Scholz A, Holtgraewe D, Winter H, Huber SC. Plant J. 2003
- ↑ Glycosyltransferases: structures, functions, and mechanisms. Lairson LL, Henrissat B, Davies GJ, Withers SG. Annu Rev Biochem. 2008