Sandbox Reserved 1128
From Proteopedia
| Line 23: | Line 23: | ||
<div class="center" style="width: auto; margin-left: auto; margin-right: auto;">[[Image:Domaine.png]]</div> | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">[[Image:Domaine.png]]</div> | ||
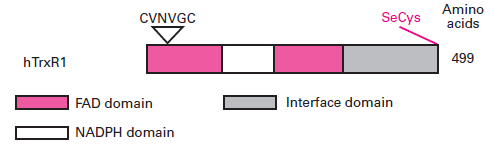
| - | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">Figure 2. Domain structure of humain TrxR1. Cited by Mustacich and Powis (2000)<ref>Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000 Feb 15;346 Pt 1:1-8.</ref> | + | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">'''Figure 2.''' Domain structure of humain TrxR1. Cited by Mustacich and Powis (2000)<ref>Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000 Feb 15;346 Pt 1:1-8.</ref> |
The catalytic-site sequences are indicated by triangles, and the position of the C-terminal penultimate SeCys residues is shown. | The catalytic-site sequences are indicated by triangles, and the position of the C-terminal penultimate SeCys residues is shown. | ||
</div> | </div> | ||
| Line 32: | Line 32: | ||
<div class="center" style="width: auto; margin-left: auto; margin-right: auto;">[[Image:Structure.png]]</div> | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">[[Image:Structure.png]]</div> | ||
| - | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">Figure 3. Model of the complex of rat TrxR with human Trx. Bound FAD (red); bound NADP (orange); interface domain (blue) and bound Trx (green). Residues at the Trx–TrxR interface are shown as stick models. The positions of the sulfur atoms of the catalytic disulfide (Cys-59–Cys-64) and the cysteine residues Cys-32, Cys-35, Cys-497, and Cys-498 are indicated by yellow spheres. Cited by Sandalova and tal. (2001).</div> | + | <div class="center" style="width: auto; margin-left: auto; margin-right: auto;">'''Figure 3.''' Model of the complex of rat TrxR with human Trx. Bound FAD (red); bound NADP (orange); interface domain (blue) and bound Trx (green). Residues at the Trx–TrxR interface are shown as stick models. The positions of the sulfur atoms of the catalytic disulfide (Cys-59–Cys-64) and the cysteine residues Cys-32, Cys-35, Cys-497, and Cys-498 are indicated by yellow spheres. Cited by Sandalova and tal. (2001).</div> |
Revision as of 18:55, 30 January 2016
Thioredoxin Reductase 1 (Human)
|
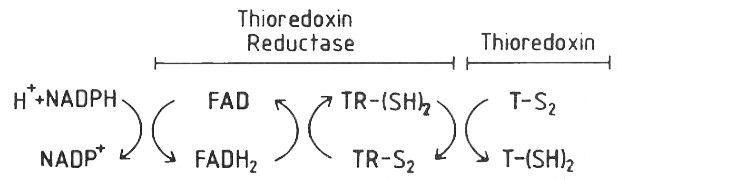
Thioredoxin reductase 1 (TrxR1) is an ubiquitous enzyme which reduces the thioredoxin protein by a disulfide oxidoreductase activity [1]. This enzyme belongs to the flavoprotein family which needs cofactors to catalyze the NADPH-dependent reaction. NADPH cofactor allows electrons transmission during the reaction via FAD from enzyme to oxidized protein. The thioredoxin system is thus composed of thioredoxin reductase, NADPH and thioredoxin. The reaction catalized is:
TrxR1 belongs to one of the two forms of mammalian TrxR enzymes mostly present in cytosol contrary to TrxR2 which is only mitochondrial. TrxR1 is a heterogeneous protein which is present in most tissues and is specific of the small thioredoxin 1 protein (Trx1) [2]. Its capacity to reduce oxidized Trx1 is important to maintain the active site of Trx1. The redox activity of reduced Trx1 is the key of its biological activity [3]. TrxR1 can thus regulate Trx1 activities by its NADPH dependent reduction specificity.
Contents |
Structure
The structure is really important to understand redox regulatory processes of the human Thioredoxin reductase 1. It could be also the template for drug design because we know that this protein is involved in various diseases.
TrxR1 is a flavoprotein belonging to pyridine nucleotide-disulfide oxido-reductase protein family which is a homopolymeric protein. The functional enzyme uses thus the flavin adenine dinucleotide (FAD) as nucleic acid cofactor. TrxR is currently the only enzyme know to reduce Trx oxidized through NADPH as the source of reducing equivalents. Function of this enzyme is correlated with structural researches.
Indeed, it has been well characterized that each monomer contains thus a FAD prosthetic group, an NADPH binding site and an active site containing a redox active disulphide which acts as proton acceptor. The active site of the human TrxR1 has been also identified as a seleneylsulfide and this selenolthiol motif is formed from a sequence conserved: Cys-Val-Asn-Val-Gly-Cys [4]. This protein has a high molecular weight: each monomer has a molecular mass of 54,6 kDa.
The catalytic-site sequences are indicated by triangles, and the position of the C-terminal penultimate SeCys residues is shown.
The results reported by Sandalova and tal. (2001) showed the crystal structure analysis of the SeCys498Cys mutant of rat TrxR complexed with NADP+ [6]. That allowed visualizing the architecture of the active site in an overall fold of the enzyme.
Each binding domain have a central five-stranded parallel β-sheet and three-stranded β-meander. In the other side of parallel sheet there are several α-helices. The two cysteins Cys59 and Cys64 which form the active disulfide, are located on helix α2. The interaction of the two subunits takes place at the interface domain which contains antiparallele five-stranded β-sheet E flanked on both sides by four helices. The C terminal domain contains an extension with a selenocysteine (Se) residue with specific motif of Gly-Cys-SeCys-Gly. The first three residues of the extension continue in the direction toward the surface of the molecule.
Catalytic mechanism
The FAD domain which contain the active site (catalytic site) and the binding site of NADPH are close to the ring of FAD. That allows the electrons to go from NADPH to the substrate through the ring of FAD and the disulphide active site. There is no big conformational change in the enzyme to do that. When there is an excess of NADPH, hTrxR forms a stable charge transfer complex.
In human TrxR there is an additional redox-active site at the C-terminal part of the chain which is not part of the conserved active site. This other redox center is characterized by the selenocysteine residue with a Cys-SeCys peptide motif which allows extension of the electron transfer chain through enzyme active site. However, it had been show that the Sec is really important and essential for the catalytic efficiency [7].
The action mechanism of Se-containing TrxR is supposed to be like that:
- • The NADPH fixes to the TrxR NADPH binding site and the electrons go from NADPH to the conserved catalytic site through FAD to reduce the disulphide bond of TrxR. So at this step the catalytic site is in its oxidized form.
- • Then there is the oxidation of the Cys-X-X-Cys site and the reduction of the C-terminal Se-containing site thanks to a Thiol-Disulfide exchange.
- • The reduced C-terminal site goes away transferring electrons to a substrate.
One suggestion for the catalytic mechanism of human TrxR is that the C-terminal conserved site is flexible which allows the Se-containing part to bring the reducing equivalents to the substrate.
Biological function
Biological functions of TrxR1 depend directly on thioredoxin (Trx) activities [8]. Trx can interact with many partners in different cellular compartments. Its biological function is cellular localisation-dependent. Reductase activity of Trx1 can regulate cell growth or apoptosis for example, but in the nucleus, Trx can bind to different transcription factors. The key role of Trx is its capacity to defence against oxidative damage.
A central role for oxidative stress
Oxidative stress is the imbalance between oxidative and reducing species and several mediators can alternate this redox potential. Most of the time, pro-oxidative species derived from di-oxygen (O2) or nitrogen monoxide (NO) in high concentration cause oxidative damages. Reactive oxygen species (ROS) have signaling functions and their increased levels are stimuli for cells. In response, cells engage into a program to change their characteristics, such as differentiation or apoptosis.
Reductase activity of Trx, activated by TrxR1, can neutralize the ROS in order to equilibrate the oxidative drift by mediating the reduction of proteins involved in scavenging ROS. [9]. Trx1 is an important hydrogen donor to ribonucleotide reductase which has an intracellular antioxidant activity. Trx acts thus as an important regulator of the oxidative stress.
Regulation of signal transduction pathways
Some cellular pathways are affected by increase of the levels of oxidizing species below those inducing damage. Several of these pathways rely on transcriptional responses by activation of redox-sensitive transcription factors such as p53, AP-1 or NF-κB in cytoplasm [10] [11]. These transcription factors are then activated by Trx in nucleus. This enzyme over-expressed can thus bind redox-sensitive transcription factors and activates them. That leads to modulate their DNA-binding activity on the promoter region of several genes. Transcription factors regulate in this way expression of genes which leads to cellular activation and regulates apoptosis [12]. For example, the tumour suppressor protein p53 stimulates reporter gene expression involved in cellular function such as mitosis or apoptosis. He is the guardian of the genome in prevent mutations by inducing expression of various genes as redox related genes, apoptosis related genes and many other [13]. In addition, Trx can also regulate the transcription factor NF-κB which is involved in the control of several processes as cell growth, immune response or even inflammation [14].
Impact on the immune system
Thioredoxin was first identified as a cytokine like factor in virus transformed cells [15]. Indeed, the Trx protein allows reduce NF-κB by its binding to this transcription factor. NF-κB factor can be thus a redox sensitive factor by regulation of gene expression of cytokines or other immune response genes. Trx allows directly regulation of pro-inflammatory cytokines expression and demonstrates several anti-inflammatory effects [16].
Diseases
Cancer
Studies showed that Thioredoxine reducatase 1 (TrxR1) is overexpressed (10 times than the normal)[17]. in many cancer cells. Even if the direct link between TrxR1 and cancer induction is not clear, there is evidence that TrxR1 regulates DNA replication[18]. Reduction in TrxR1 activity results in blockage of cells in phase S. Indeed, decrease in TrxR1 concentration leads to decrease in reduced-Trx (activated form) which normally acts as an electron donor for the Ribonucleotide reductase (the enzyme that syntehises most deoxyribonucleotides from ribonucleotides). Some TrxR1 isoforms are thought to guide actin and tubulin polymerization[19]. Inactive-form Trx are also unable to inhibit apoptosis, as unable to inhibit the Apoptosis Signal-regulating Kinase 1 (ASK1), also known as Mitogen-Activated Protein Kinase Kinase Kinase 5 (MAP3K5). Furthermore, a studie showed that the Thioredoxin system, cooperating with the redox factor-1 (APE/Ref-1) regulates basal activity of p53 [20]. TrxR1 is then a good target for cancer treatement. Recent studies showed that arsenic trioxide AS2O3 or ATO inhibits TrxR1[21] in its redox state in presence of NADPH and is an approved anti-cancer chemotherapeutic drug commercially named Trisenox®[22]. It is used to treat acute promyelocytic leukemia, multiple myeloma, chronic myelogenous leukemia, and acute myelogenous leukemia.
Cardiovascular diseases
Thioredoxin Reductase 1 (TrxR1) has important protection functions against oxidative stress due to its antioxidant properties. Studies showed that downregulation of TrxR1 inducing Trx downregulation leads to cardiomyocyte injuries. TrxR, indirectly via TrX, plays a protective role in myocardial ischemia/reperfusion[23]. TrxR1, part of the Thioredoxin system is suggested to play a role in cellular defense against oxidized LDL and arteriosclerosis development. TrxR could also be involved in hypertension and diabetes as anormal levels of thioredoxin is found in hypertensive and diabetic patients serum compared to normal patients[24].
Neuronal diseases
Thioredoxin Reductase 1 (TrxR1) could be related to Parkinston’s Disease as TrxR1 expression decreased in the substantia nigra pars compacta of the Parkinson's disease mouse model, suggesting that TrxR1 may play a protective antioxidant role preventing neurodegeneration[25]. Although the direct involvement of Thioredoxin Reductase 1 is not clear in Alzheimer disease, studies showed that patients diagnosed with Alzheimer have a higher level of Thioredoxin Reductase 1 (TrxR1) in their brain[26].
AIDS and inflamatory diseases
Thioredoxin Reductase 1 (TrxR1) plays an important role in repressing HIV-1 protease by negatively regulating the HIV-1 transcriptional activator in human macrophages[27]. Studies suggested that TrxR1, by regulating ROS generation, regulate the redox-dependent signal transduction and may confer protection against inflammatory processes induced by influenza virus[28].
</StructureSection>
References
- ↑ Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000 Feb 15;346 Pt 1:1-8.
- ↑ Jurado J, Prieto-Alamo MJ, Madrid-Rísquez J, Pueyo C. Absolute gene expression patterns of thioredoxin and glutaredoxin redox systems in mouse. J Biol Chem. 2003 Nov 14;278(46):45546-54. Epub 2003 Sep 3.
- ↑ Holmgren A, Björnstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199-208.
- ↑ Lothrop AP, Snider GW, Ruggles EL, Patel AS, Lees WJ, Hondal RJ. Selenium as an electron acceptor during the catalytic mechanism of thioredoxin reductase. Biochemistry. 2014 Feb 4;53(4):654-63. doi: 10.1021/bi400658g. Epub 2014 Jan 23.
- ↑ Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000 Feb 15;346 Pt 1:1-8.
- ↑ Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9533-8. Epub 2001 Jul 31.
- ↑ Lee SR, Bar-Noy S, Kwon J, Levine RL, Stadtman TC, Rhee SG. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc Natl Acad Sci U S A. 2000 Mar 14;97(6):2521-6.
- ↑ Oblong JE, Berggren M, Gasdaska PY, Powis G. Site-directed mutagenesis of active site cysteines in human thioredoxin produces competitive inhibitors of human thioredoxin reductase and elimination of mitogenic properties of thioredoxin. J Biol Chem. 1994 Apr 22;269(16):11714-20.
- ↑ Powis G, Montfort WR. Properties and biological activities of thioredoxins. Annu Rev Pharmacol Toxicol. 2001;41:261-95.
- ↑ Ueno M, Masutani H, Arai RJ, Yamauchi A, Hirota K, Sakai T, Inamoto T, Yamaoka Y, Yodoi J, Nikaido T. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem. 1999 Dec 10;274(50):35809-15.
- ↑ Freemerman AJ, Gallegos A, Powis G. Nuclear factor kappaB transactivation is increased but is not involved in the proliferative effects of thioredoxin overexpression in MCF-7 breast cancer cells. Cancer Res. 1999 Aug 15;59(16):4090-4.
- ↑ Cassidy PB, Edes K, Nelson CC, Parsawar K, Fitzpatrick FA, Moos PJ. Thioredoxin reductase is required for the inactivation of tumor suppressor p53 and for apoptosis induced by endogenous electrophiles. Carcinogenesis. 2006 Dec;27(12):2538-49. Epub 2006 Jun 15.
- ↑ Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997 Sep 18;389(6648):300-5.
- ↑ Kabe Y, Ando K, Hirao S, Yoshida M, Handa H. Redox regulation of NF-kappaB activation: distinct redox regulation between the cytoplasm and the nucleus. Antioxid Redox Signal. 2005 Mar-Apr;7(3-4):395-403.
- ↑ Nakamura H. Extracellular functions of thioredoxin. Novartis Found Symp. 2008;291:184-92; discussion 192-5, 221-4.
- ↑ Bertini R, Howard OM, Dong HF, Oppenheim JJ, Bizzarri C, Sergi R, Caselli G, Pagliei S, Romines B, Wilshire JA, Mengozzi M, Nakamura H, Yodoi J, Pekkari K, Gurunath R, Holmgren A, Herzenberg LA, Herzenberg LA, Ghezzi P. Thioredoxin, a redox enzyme released in infection and inflammation, is a unique chemoattractant for neutrophils, monocytes, and T cells. J Exp Med. 1999 Jun 7;189(11):1783-9.
- ↑ Berggren M, Gallegos A, Gasdaska JR, Gasdaska PY, Warneke J, Powis G. Thioredoxin and thioredoxin reductase gene expression in human tumors and cell lines, and the effects of serum stimulation and hypoxia. Anticancer Res. 1996 Nov-Dec;16(6B):3459-66.
- ↑ Min-Hyuk Yoo, Xue-Ming Xu, Bradley A. Carlson, Andrew D. Patterson, Vadim N. Gladyshev, and Dolph L. Hatfield. Targeting Thioredoxin Reductase 1 Reduction in Cancer Cells Inhibits Self-Sufficient Growth and DNA Replication. PLoS ONE. 2007; 2(10): e1112.
- ↑ Dammeyer P, Damdimopoulos AE, Nordman T, Jiménez A, Miranda-Vizuete A, Arnér ES. Induction of cell membrane protrusions by the N-terminal glutaredoxin domain of a rare splice variant of human thioredoxin reductase 1. J Biol Chem. 2008 Feb 1;283(5):2814-21. Epub 2007 Nov 27.
- ↑ Séverine Seemann and Pierre Hainaut. Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene (2005) 24, 3853–3863. doi:10.1038
- ↑ Lu J, Chew EH, Holmgren A. Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc Natl Acad Sci U S A. 2007 Jul 24;104(30):12288-93. Epub 2007 Jul 18
- ↑ Arsenic Trioxide, Chemocare(chemocare.com). Copyright © 2002 - 2016 by Chemocare.com ® All rights reserved.
- ↑ Ashok K. Srivastava, Madhu B. Anand-Srivastava. Signal Transduction in the Cardiovascular System in Health and Disease. Springer Science & Business Media. 2008 sept 20; pp145-147
- ↑ Ashok K. Srivastava, Madhu B. Anand-Srivastava. Signal Transduction in the Cardiovascular System in Health and Disease. Springer Science & Business Media. 2008 sept 20; pp147-148
- ↑ Zihua Liu, Yuhong Jing, Jie Yin, Jiying Mu, Tingting Yao, and Liping Gao, Ph.D. Downregulation of thioredoxin reductase 1 expression in the substantia nigra pars compacta of Parkinson's disease mice. Neural Regen Res. 2013 Dec 15; 8(35): 3275–3283.
- ↑ Lovell MA, Xie C, Gabbita SP, Markesbery WR. Decreased thioredoxin and increased thioredoxin reductase levels in Alzheimer's disease brain. Free Radic Biol Med. 2000 Feb 1;28(3):418-27
- ↑ Kalantari P, Narayan V, Natarajan SK, Muralidhar K, Gandhi UH, Vunta H, Henderson AJ, Prabhu KS. Thioredoxin reductase-1 negatively regulates HIV-1 transactivating protein Tat-dependent transcription in human macrophages. J Biol Chem. 2008 Nov 28;283(48):33183-90.
- ↑ Hirokazu Tsukahara, Kazunari Kaneko. Studies on Pediatric Disorders. Springer 2014 mai 23; pp 233-236.