This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
1e2h
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1e2h.gif|left|200px]] | [[Image:1e2h.gif|left|200px]] | ||
| - | + | <!-- | |
| - | + | The line below this paragraph, containing "STRUCTURE_1e2h", creates the "Structure Box" on the page. | |
| - | + | You may change the PDB parameter (which sets the PDB file loaded into the applet) | |
| - | + | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | |
| - | + | or leave the SCENE parameter empty for the default display. | |
| - | | | + | --> |
| - | | | + | {{STRUCTURE_1e2h| PDB=1e2h | SCENE= }} |
| - | + | ||
| - | + | ||
| - | }} | + | |
'''THE NUCLEOSIDE BINDING SITE OF HERPES SIMPLEX TYPE 1 THYMIDINE KINASE ANALYZED BY X-RAY CRYSTALLOGRAPHY''' | '''THE NUCLEOSIDE BINDING SITE OF HERPES SIMPLEX TYPE 1 THYMIDINE KINASE ANALYZED BY X-RAY CRYSTALLOGRAPHY''' | ||
| Line 29: | Line 26: | ||
[[Category: Schulz, G E.]] | [[Category: Schulz, G E.]] | ||
[[Category: Vogt, J.]] | [[Category: Vogt, J.]] | ||
| - | [[Category: | + | [[Category: Adenine analog]] |
| - | [[Category: | + | [[Category: Enzyme-prodrug gene therapy]] |
| - | [[Category: | + | [[Category: Nucleoside-binding]] |
| - | [[Category: | + | [[Category: Thymidine kinase]] |
| - | [[Category: | + | [[Category: X-ray crystallography]] |
| - | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Fri May 2 14:34:37 2008'' | |
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | |
Revision as of 11:34, 2 May 2008
| |||||||||
| 1e2h, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Activity: | Thymidine kinase, with EC number 2.7.1.21 | ||||||||
| Related: | 1kim, 1vtk, 2vtk, 3vtk, 1ki2, 1ki4, 1ki5, 1ki6, 1ki7, 1ki8, 1e2i, 1e2j, 1e2k, 1e2l, 1e2m, 1e2n, 1e2p | ||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
THE NUCLEOSIDE BINDING SITE OF HERPES SIMPLEX TYPE 1 THYMIDINE KINASE ANALYZED BY X-RAY CRYSTALLOGRAPHY
Overview
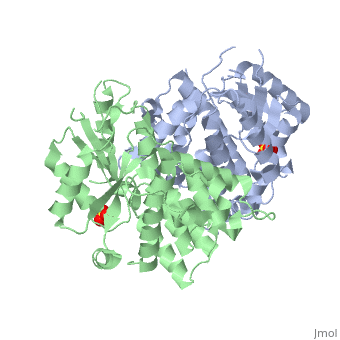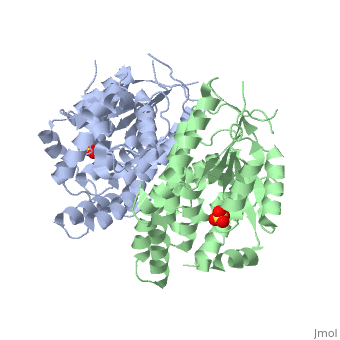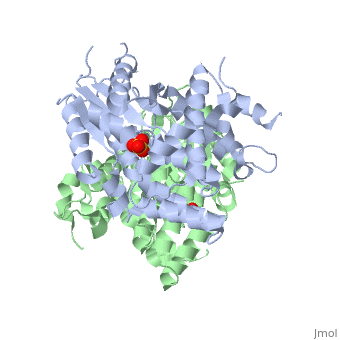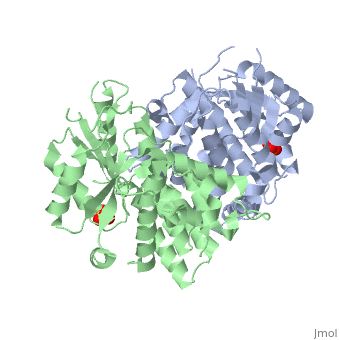
The crystal structures of the full-length Herpes simplex virus type 1 thymidine kinase in its unligated form and in a complex with an adenine analogue have been determined at 1.9 A resolution. The unligated enzyme contains four water molecules in the thymidine pocket and reveals a small induced fit on substrate binding. The structure of the ligated enzyme shows for the first time a bound adenine analogue after numerous complexes with thymine and guanine analogues have been reported. The adenine analogue constitutes a new lead compound for enzyme-prodrug gene therapy. In addition, the structure of mutant Q125N modifying the binding site of the natural substrate thymidine in complex with this substrate has been established at 2.5 A resolution. It reveals that neither the binding mode of thymidine nor the polypeptide backbone conformation is altered, except that the two major hydrogen bonds to thymidine are replaced by a single water-mediated hydrogen bond, which improves the relative acceptance of the prodrugs aciclovir and ganciclovir compared with the natural substrate. Accordingly, the mutant structure represents a first step toward improving the virus-directed enzyme-prodrug gene therapy by enzyme engineering.
About this Structure
1E2H is a Single protein structure of sequence from Human herpesvirus 1. Full crystallographic information is available from OCA.
Reference
Nucleoside binding site of herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography., Vogt J, Perozzo R, Pautsch A, Prota A, Schelling P, Pilger B, Folkers G, Scapozza L, Schulz GE, Proteins. 2000 Dec 1;41(4):545-53. PMID:11056041 Page seeded by OCA on Fri May 2 14:34:37 2008