We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
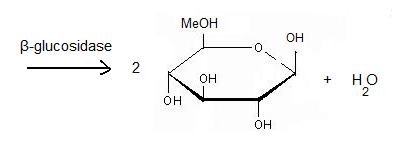
Beta-glucosidase
From Proteopedia
(Difference between revisions)
| Line 54: | Line 54: | ||
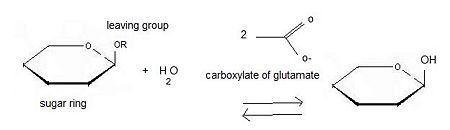
The second step: '''hydrolysis''' is the most important. A cellulase is a complex of 3 enzymes which act together to hydrolyse cellulose: Endoglucanase breaks the chain in the middle of the molecular structure of cellulose. Exoglucanase binds an available end of the chain and isolates it. Then units of cellobiose are cut (two units of glucose which are together). To finish, '''β-glucosidase''' divides cellobiose into two glucoses. When they ferment, they become ethanol. The final product is obtained thanks to fermentation, distillation and deshydratation. | The second step: '''hydrolysis''' is the most important. A cellulase is a complex of 3 enzymes which act together to hydrolyse cellulose: Endoglucanase breaks the chain in the middle of the molecular structure of cellulose. Exoglucanase binds an available end of the chain and isolates it. Then units of cellobiose are cut (two units of glucose which are together). To finish, '''β-glucosidase''' divides cellobiose into two glucoses. When they ferment, they become ethanol. The final product is obtained thanks to fermentation, distillation and deshydratation. | ||
| + | == Human acid-beta-glucosidase covalently bound to conduritol B epoxide == | ||
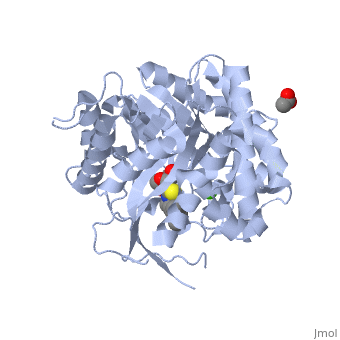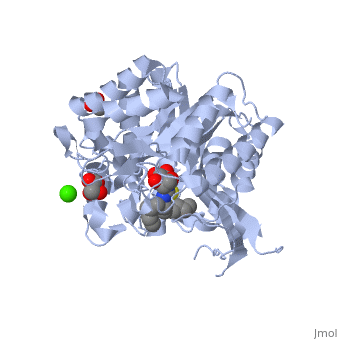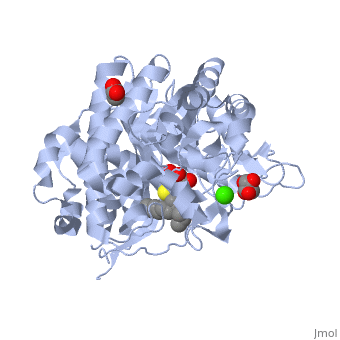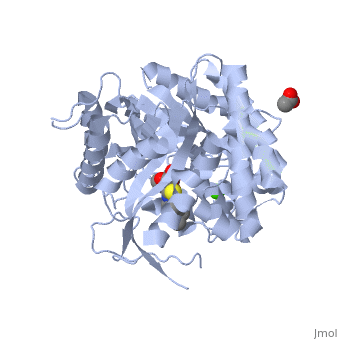
| + | The [[X-ray crystallography|crystal structure]] of the human acid-β-glucosidase ([[acid-beta-glucosidase]], [http://en.wikipedia.org/wiki/Glucocerebrosidase glucocerebrosidase], <scene name='1y7v/Active_site/12'>GlcCerase</scene>, [http://www.expasy.org/cgi-bin/nicezyme.pl?3.2.1.45 E.C. 3.2.1.45], <span style="color:yellow;background-color:black;font-weight:bold;">colored yellow</span> with covalently bound [http://en.wikipedia.org/wiki/Enzyme_inhibitor#Irreversible_inhibitors irreversible inhibitor] <scene name='1y7v/Bound_cyclohexitol/4'>cyclohexitol</scene> (<span style="color:cyan;background-color:black;font-weight:bold;">conduritol-B-epoxide; CBE; shown in cyan</span> with its <font color='red'><b>hydroxyl groups</b></font> are in <font color='red'><b>red</b></font>) was solved. This structure reveals that binding of CBE to the [http://en.wikipedia.org/wiki/Active_site active site] does not induce a global conformational change in GlcCerase and confirms that Glu340 is the active-site catalytic [http://en.wikipedia.org/wiki/Nucleophile nucleophile], because the <scene name='1y7v/Active_site1/3'>distance</scene> between the cyclohexitol C1 atom and Glu340 Oε2 is 1.43 Å. The comparison between the active sites of <scene name='1y7v/Active_site/13'>GlcCerase</scene> and another representative of the glycohydrolase family - plant <scene name='1y7v/Active_site_beta_glu_glyco/5'>β-D-glucan glucohydrolase</scene> ([[1iev]]), reveals that CBE bound with this plant enzyme adopted the "chair" conformation, while with human <scene name='1y7v/Active_site/14'>GlcCerase</scene>, it is observed in a "boat" conformation, with hydrogen bonds to Asn234 Oδ1 and Nδ2, Glu340 Oε1, Trp179 Nε1, and Asp127 Oδ1 and Oδ2. | ||
| + | {{Clear}} | ||
| + | |||
| + | Only one of two <scene name='1y7v/Loops/3'>alternative conformations</scene> of a pair of flexible loops (L1: Ser345–Glu349, and L2: Val394–Asp399) located at the entrance to the active site in native GlcCerase ([[1ogs]]) is observed in the GlcCerase-CBE structure, a conformation in which the active site is accessible to CBE (<font color='blue'><b>colored blue</b></font>), while these loops in <font color='magenta'><b>the second (closed) conformation are colored magenta</b></font>. In <scene name='1y7v/L2/5'>loop 2</scene>, a major structural change is observed in the positions of <scene name='1y7v/L2/6'>Asn396 and Phe397</scene>, and in <scene name='1y7v/L1/6'>loop 1</scene> a more limited difference is observed in the conformations of <scene name='1y7v/L1/7'>Lys346 and Glu349</scene>. Analysis of the dynamics of these two alternative conformations suggests that the two loops act as a lid at the entrance to the active site. The movies [http://www.jbc.org/content/vol0/issue2005/images/data/M502799200/DC1/mov.mov 1] and [http://www.jbc.org/content/vol0/issue2005/images/data/M502799200/DC1/mov2.mov 2] illustrate the dynamics of the movement of these two loops. | ||
==Additional Resources== | ==Additional Resources== | ||
For additional information, see: [[Carbohydrate Metabolism]] | For additional information, see: [[Carbohydrate Metabolism]] | ||
Revision as of 08:16, 9 February 2016
| |||||||||||
3D structures of Beta-glucosidase
Updated on 09-February-2016
References
- ↑ Aguilar M, Gloster TM, Garcia-Moreno MI, Ortiz Mellet C, Davies GJ, Llebaria A, Casas J, Egido-Gabas M, Garcia Fernandez JM. Molecular basis for beta-glucosidase inhibition by ring-modified calystegine analogues. Chembiochem. 2008 Nov 3;9(16):2612-8. PMID:18833549 doi:10.1002/cbic.200800451
- ↑ http://en.wikipedia.org/wiki/B-glucosidase
- ↑ Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853-9. PMID:8535779
- ↑ http://www.ebi.ac.uk/interpro/IEntry?ac=IPR018120#PUB00002205
- ↑ http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/CSA/CSA_Site_Wrapper.pl?pdb=2vrj
- ↑ Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853-9. PMID:8535779
- ↑ http://www.cazy.org/fam/ghf_INV_RET.html#3
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Muriel Breteau, Alexander Berchansky, Joel L. Sussman, David Canner