We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 439
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
| - | |||
| - | =='''Asp Receptor Ligand-binding domain (1wat)<ref>PMID: 8486661</ref>'''== | ||
<StructureSection load='1wat' size='350' side='right' caption='Bacteria use this protein to "smell" their environment (PDB entry [[1wat]])' scene=''> | <StructureSection load='1wat' size='350' side='right' caption='Bacteria use this protein to "smell" their environment (PDB entry [[1wat]])' scene=''> | ||
| + | =='''Asp Receptor Ligand-binding domain (1wat)<ref>PMID: 8486661</ref>'''== | ||
| + | |||
by [list all teammate names here] | by [list all teammate names here] | ||
| Line 13: | Line 13: | ||
<font color='orange'>This is an example page. Edit appropriately for your project. Upon completion of each task, delete instructions in orange. See [[Sandbox Reserved 439]] for original instructions</font> | <font color='orange'>This is an example page. Edit appropriately for your project. Upon completion of each task, delete instructions in orange. See [[Sandbox Reserved 439]] for original instructions</font> | ||
| - | <font color='orange'>1. Add a citation to the primary reference at the end of the title above: you should all read this paper! | + | <font color='orange'>1. Add a citation to the primary reference at the end of the title above: you should all read this paper! Go to the [http://www.rcsb.org/pdb/home/home.do pdb] and search for your pdb code. Go down to the abstract and click "Search on Pubmed". |
| - | Go to the [http://www.rcsb.org/pdb/home/home.do pdb] and search for your pdb code. | + | |
| - | Go down to the abstract and click "Search on Pubmed". | + | |
In pubmed, copy the PMID code you see right below the abstract, and paste it to replace xxx after PMID in your title (first edited line above).</font> | In pubmed, copy the PMID code you see right below the abstract, and paste it to replace xxx after PMID in your title (first edited line above).</font> | ||
| Line 25: | Line 23: | ||
==Binding Interactions== | ==Binding Interactions== | ||
| + | <font color='orange'>Each section should start with an "initial view" green scene. These can be the same for all sections or can be set by the section author.</font> | ||
<scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>Initial view</scene> | <scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>Initial view</scene> | ||
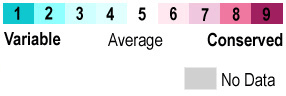
When the protein is colored according to <scene name='69/695684/Conservation/1'>sequence conservation</scene> , residues at the ligand site are the most conserved. | When the protein is colored according to <scene name='69/695684/Conservation/1'>sequence conservation</scene> , residues at the ligand site are the most conserved. | ||
| Line 32: | Line 31: | ||
==Additional Features== | ==Additional Features== | ||
| + | <font color='orange'>Each section should start with an "initial view" green scene. These can be the same for all sections or can be set by the section author.</font> | ||
<scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>Initial view</scene> | <scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>Initial view</scene> | ||
==Quiz Question 1== | ==Quiz Question 1== | ||
| + | <font color='orange'>Each section should start with an "initial view" green scene. These can be the same for all sections or can be set by the section author.</font> | ||
<scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>Initial view</scene> | <scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>Initial view</scene> | ||
Revision as of 18:38, 2 March 2016
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
| |||||||||||