We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Daniel Schemenauer/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
== Introduction == | == Introduction == | ||
=== Biological Background === | === Biological Background === | ||
| - | G-Coupled Protein Receptors (GPCRs) are divided into four major classes; Class A,B,C, and F.<ref name="MSGPCR"/> | + | G-Coupled Protein Receptors (GPCRs) are divided into four major classes; Class A,B,C, and F.<ref name="MSGPCR">PMID:23407534</ref> |
=== Importance === | === Importance === | ||
| - | + | <ref name="1MT5">PMID:12459591</ref> | |
== Structure == | == Structure == | ||
| Line 30: | Line 30: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | <ref>PMID:23407534</ref | + | <ref>PMID:23407534</ref> |
This is a default text for your page '''Daniel Schemenauer/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Daniel Schemenauer/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI | ||
10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
{{reflist}} | {{reflist}} | ||
Revision as of 13:21, 22 March 2016
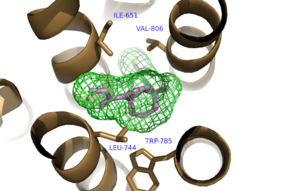
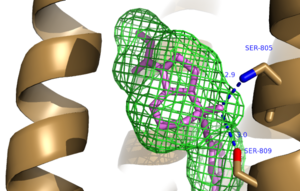
| |||||||||||
References
- ↑ Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013 Feb 14;494(7436):185-94. doi: 10.1038/nature11896. PMID:23407534 doi:http://dx.doi.org/10.1038/nature11896
- ↑ Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002 Nov 29;298(5599):1793-6. PMID:12459591 doi:10.1126/science.1076535
[1] This is a default text for your page Daniel Schemenauer/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [2] or to the article describing Jmol [3] to the rescue.
- ↑ Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013 Feb 14;494(7436):185-94. doi: 10.1038/nature11896. PMID:23407534 doi:http://dx.doi.org/10.1038/nature11896
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644