We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Ke Xiao/Geobacter pilus models
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
===Pilus Model=== | ===Pilus Model=== | ||
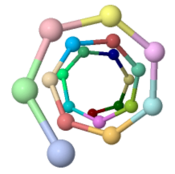
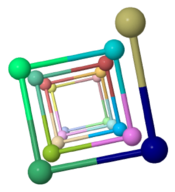
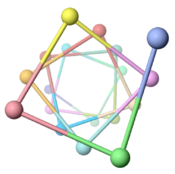
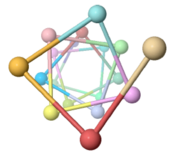
| - | The theoretical ''Geobacter sulfurreducens'' pilus model shown here, called ''ARC-1''<ref name="xiao1"> | + | The theoretical ''Geobacter sulfurreducens'' pilus model shown here, called ''ARC-1''<ref name="xiao1">PMID: 27001169</ref> (<scene name='69/699340/Arc-1_no_hydrogen/1'>restore initial scene</scene>), is representative of a cluster of 50 low-energy models with an arrangement of aromatic rings consistent with X-ray diffraction data<ref name="malvankar2015">PMID: 25736881</ref>. Unlike the docking of crystallographic models into electron density from cryo-electron microscopy<ref>PMID: 16949362</ref><ref>PMID: 12769840</ref>, these models have chemically realistic interactions between subunit chains. Energy minimization produced subunit interactions with shape complementarity, non-covalent bonds, and very few atomic clashes<ref>The chemical realism of these theoretical energy-minimized models contrasts with models where empirical monomer structures are docked into cryo-electron microscopic electron density maps. Those are unrealistic in details of subunit interactions, lacking shape complementarity and having many atomic clashes. See "Initial Model Outputs" in [http://www.nature.com/articles/srep23385 the publication] for details.</ref>. |
===Monomer Chains=== | ===Monomer Chains=== | ||
Revision as of 20:50, 28 March 2016
Interactive 3D Complement in Proteopedia
Scientific Reports an online, open access journal: nature.com/srep
Low energy atomic models suggesting a pilus structure that could account for electrical conductivity of Geobacter sulfurreducens pili.
Ke Xiao, Nikhil S. Malvankar, Chuanjun Shu, Eric Martz, Derek R. Lovley, and Xiao Sun.
Scientific Reports 6:23385, March 2016: nature.com/articles/srep23385. (DOI: 10.1038/srep23385)
Molecular Tour
| |||||||||||
| Theoretical Model: The protein structure described on this page was determined theoretically, and hence should be interpreted with caution. |
Download
Pilus Model
- Click to download Geobacter sulfurreducens pilus model ARC-1
Animations for Powerpoint
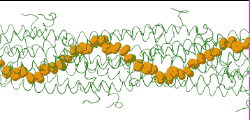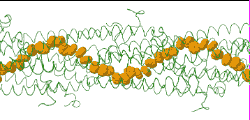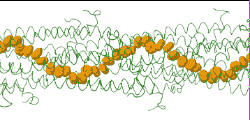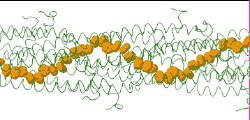
| Low resolution, ARC-1 model (smoothed green backbone traces) with aromatic rings of residues Phe1, Phe24, and Tyr27 in orange. DOWNLOAD HIGH RESOLUTION ANIMATION (19 MB). |

| Low resolution, ARC-1 model with spacefilling (van der Waals) atoms, each chain a different color. DOWNLOAD HIGH RESOLUTION ANIMATION (29 MB). |

| Low resolution, ARC-1 model with translucent spacefilling atoms. Aromatic rings of Phe1, Phe24, and Tyr27 are opaque. Each chain is a different color. DOWNLOAD HIGH RESOLUTION ANIMATION (28 MB). |
See Also
Notes & References
- ↑ 1.0 1.1 1.2 Xiao K, Malvankar NS, Shu C, Martz E, Lovley DR, Sun X. Low Energy Atomic Models Suggesting a Pilus Structure that could Account for Electrical Conductivity of Geobacter sulfurreducens Pili. Sci Rep. 2016 Mar 22;6:23385. doi: 10.1038/srep23385. PMID:27001169 doi:http://dx.doi.org/10.1038/srep23385
- ↑ 2.0 2.1 Malvankar NS, Vargas M, Nevin K, Tremblay PL, Evans-Lutterodt K, Nykypanchuk D, Martz E, Tuominen MT, Lovley DR. Structural basis for metallic-like conductivity in microbial nanowires. MBio. 2015 Mar 3;6(2):e00084. doi: 10.1128/mBio.00084-15. PMID:25736881 doi:http://dx.doi.org/10.1128/mBio.00084-15
- ↑ Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006 Sep 1;23(5):651-62. PMID:16949362 doi:10.1016/j.molcel.2006.07.004
- ↑ Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, Forest KT, Tainer JA. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003 May;11(5):1139-50. PMID:12769840
- ↑ The chemical realism of these theoretical energy-minimized models contrasts with models where empirical monomer structures are docked into cryo-electron microscopic electron density maps. Those are unrealistic in details of subunit interactions, lacking shape complementarity and having many atomic clashes. See "Initial Model Outputs" in the publication for details.
- ↑ Each chain contains the 61 C-terminal amino acids of UniProt Q74D23.
- ↑ Campos M, Francetic O, Nilges M. Modeling pilus structures from sparse data. J Struct Biol. 2011 Mar;173(3):436-44. doi: 10.1016/j.jsb.2010.11.015. Epub 2010 , Nov 27. PMID:21115127 doi:http://dx.doi.org/10.1016/j.jsb.2010.11.015
- ↑ Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004 May;2(5):363-78. PMID:15100690 doi:http://dx.doi.org/10.1038/nrmicro885
- ↑ Li J, Egelman EH, Craig L. Structure of the Vibrio cholerae Type IVb Pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. J Mol Biol. 2012 Apr 20;418(1-2):47-64. doi: 10.1016/j.jmb.2012.02.017. Epub 2012, Feb 21. PMID:22361030 doi:http://dx.doi.org/10.1016/j.jmb.2012.02.017
- ↑ Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006 Sep 1;23(5):651-62. PMID:16949362 doi:10.1016/j.molcel.2006.07.004