User:Dean Williams/Sandbox 1180
From Proteopedia
| Line 90: | Line 90: | ||
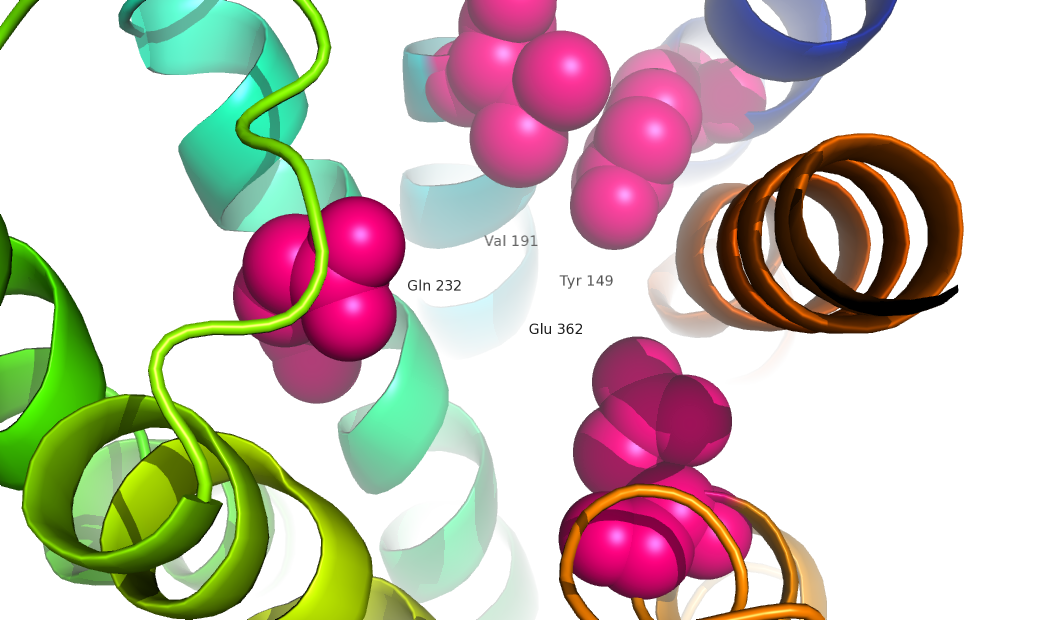
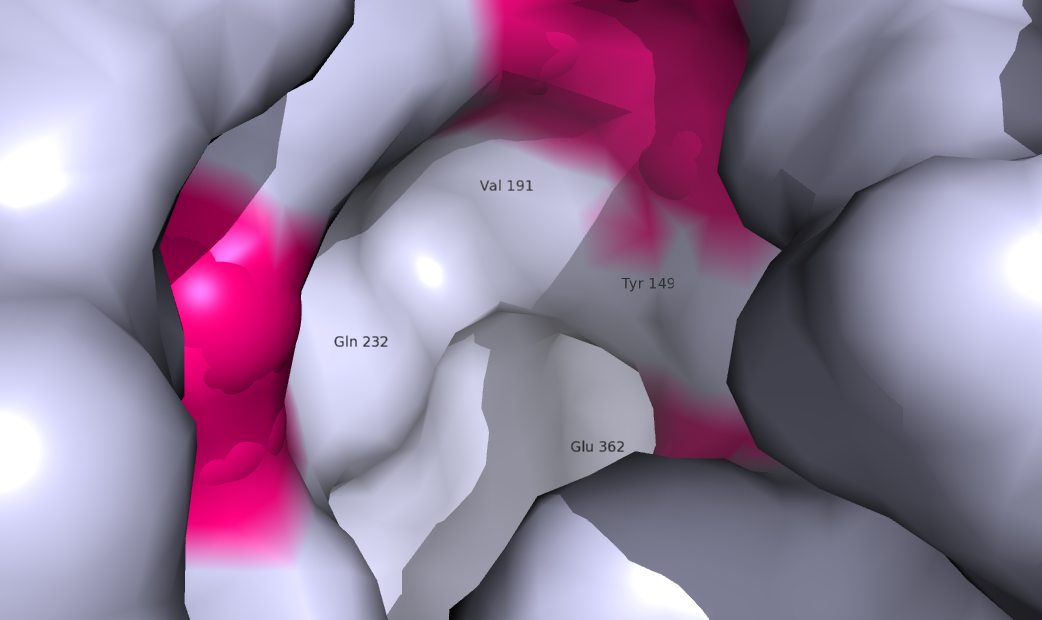
===Active binding domains/sites=== | ===Active binding domains/sites=== | ||
| - | [[Image:Movie Frame 8.png]][[Image:Movie Frame 3.png]][[Image:Movie Frame | + | [[Image:Movie Frame 8.png]][[Image:Movie Frame 3.png]][[Image:Movie Frame 6.png]][[Image:Glucagon with Q3 and N-terminus.png]] |
===Biological function of glucagon=== | ===Biological function of glucagon=== | ||
Revision as of 02:43, 30 March 2016
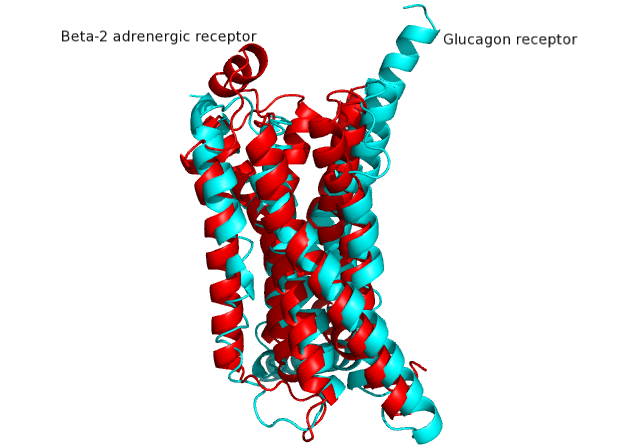
Structure of Class B Human Glucagon G-Protein Coupled Receptors
G protein coupled receptors (GPCRs) are recognized as the largest known class of integral membrane proteins and are divided into five families; the rhodopsin family (class A), the secretin family (class B), the adhesion family, the glutamate family (class C), and the frizzled/taste family (class F). Roughly 5% of the human genome encodes g-protein-coupled receptors which are responsible for the transduction of endogenous signals and the instigation of cellular response. The variants all contain a similar seven α-helical transmembrane domain (TMD) that, once bound to its peptide ligand, undergoes conformational change and tranduces a signal to coupled, heterotrimeric G proteins which initiate intracellular signal pathways and generate physiological and pathological processes. [1]
Class B GPCRs contain 15 distinct receptors for peptide hormones and generate their signal pathway through the activation of adenylate cyclase (AC) which increases concentration of cAMP, inositol phosphate, and calcium levels in cyto. [2] These signals are essential elements of intracellular signal cascades for human diseases including type II diabetes mellitus, osteoporosis, obesity, cancer, neurological degeneration, cardiovascular diseases, headaches, and psychiatric disorders; making their regulation through drug targeting of particular interest to companies developing novel molecules. [3] Structurally based approaches to the development of small-molecule agonists and antagonists have been hampered by the lack of accurate Class B TMD visualizations until recent crystal structures of corticoptropin-releasing factor receptor 1 and human glucagon were realized. [4] [5]
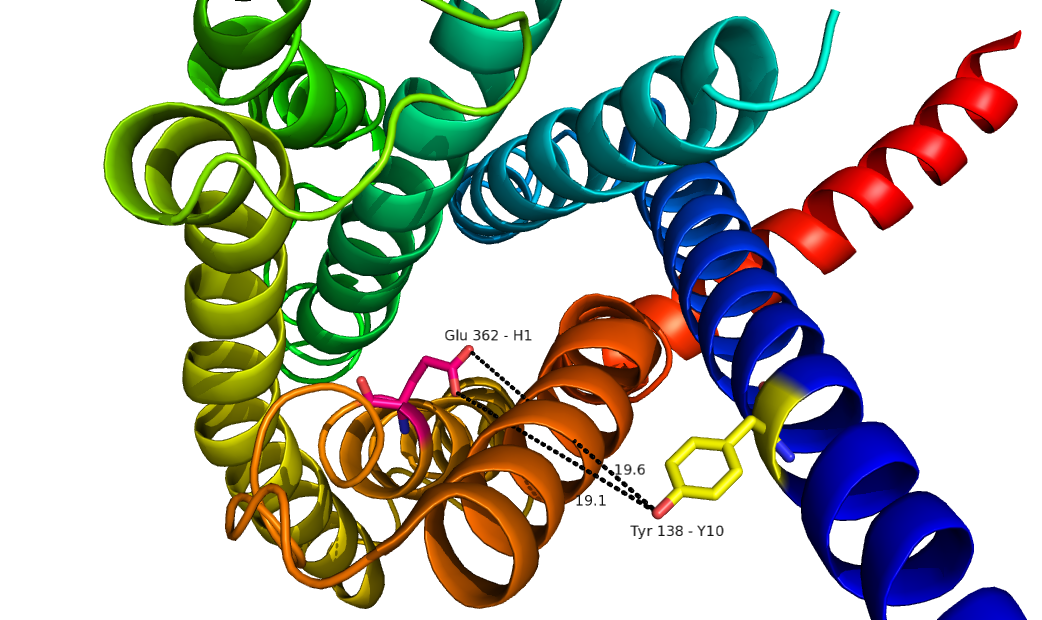
The glucagon class B GPCR is particular involved in glucose homeostasis through the binding of the signal peptide glucagon.
| |||||||||||
References
- ↑ Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006 Feb;2(2):e13. Epub 2006 Feb 17. PMID:16485037 doi:http://dx.doi.org/10.1371/journal.pcbi.0020013
- ↑ Bortolato A, Dore AS, Hollenstein K, Tehan BG, Mason JS, Marshall FH. Structure of Class B GPCRs: new horizons for drug discovery. Br J Pharmacol. 2014 Jul;171(13):3132-45. doi: 10.1111/bph.12689. PMID:24628305 doi:http://dx.doi.org/10.1111/bph.12689
- ↑ Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014 Jan;35(1):12-22. doi: 10.1016/j.tips.2013.11.001. Epub, 2013 Dec 18. PMID:24359917 doi:http://dx.doi.org/10.1016/j.tips.2013.11.001
- ↑ Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, Jazayeri A, Cooke RM, Weir M, Marshall FH. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013 Jul 25;499(7459):438-43. doi: 10.1038/nature12357. Epub 2013 Jul 17. PMID:23863939 doi:http://dx.doi.org/10.1038/nature12357
- ↑ Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644