We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1165
From Proteopedia
(Difference between revisions)
| Line 30: | Line 30: | ||
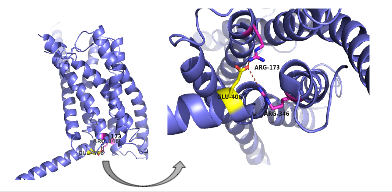
| - | There are specific amino acid interactions that maximize affinity. This includes the alpha helical structure of the <scene name='72/721535/Stalk/1'>stalk</scene>. The alpha helical structure of the stalk interacts directly with glucagon | + | There are specific amino acid interactions that maximize affinity. This includes the alpha helical structure of the <scene name='72/721535/Stalk/1'>stalk</scene>. The alpha helical structure of the stalk interacts directly with glucagon, as it extends nearly three helical turns above the membrane. When the alpha helix of the stalk is disrupted, the affinity of glucagon for GCGR decreases. A [https://en.wikipedia.org/wiki/Mutagenesis mutagenesis] study mutating <scene name='72/721535/Ala135/1'>alanine 135</scene> to a proline. Proline disrupts helices. The Ala135Pro mutant had significant lower affinity for glucagon.<ref name="Tips">PMID: 23863937</ref> Furthermore, there are certain interactions that hold the helices of the 7TM in the conformation that maximizes [http://www.chemicool.com/definition/affinity.html affinity]. <ref name="Ligands">PMID: 21542831</ref> The high affinity conformation of GCGR is the open conformation, when glucagon can bind. Without these specific interactions between the residues, the open conformation is not stabilized and GCGR remains in the closed conformation, where glucagon cannot bind. <ref name="Tips">PMID: 23863937</ref> The [https://en.wikipedia.org/wiki/Disulfide disulfide bond] between <scene name='72/721535/Disulfide_bond_notspin_actual/1'> Cys 294 and Cys 224</scene> serves to hold the helices in the proper orientation for binding and stabilizes the open conformation. Additionally, the [https://en.wikipedia.org/wiki/Salt_bridge_%28protein_and_supramolecular%29 salt bridges] between Glu 406, Arg 173, and Arg 346 hold the open conformation together for higher affinity (Figure 5). <ref name="Ligands">PMID: 21542831</ref> |
| Line 47: | Line 47: | ||
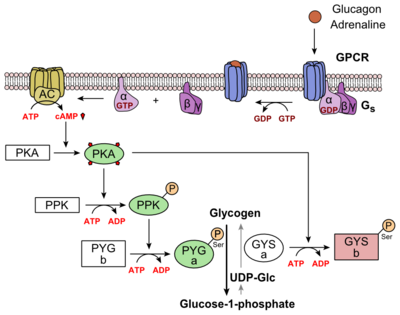
Of the fifteen human class B GPCRs, eight have been confirmed as potential [https://en.wikipedia.org/wiki/Biological_target drug target]. <ref name="Drug">PMID: 24628305</ref> However, overall family B GPCRS have been difficult drug targets. This difficulty is partially related to the inherent flexibility in the class B GCGR 7TM. The flexibility comes from the ability of GCGR to be a receptor many ligands. The ECD and it's role in interactions on the extracellular side of receptors may provide evidence to how class B receptors adjust the conformational spectra for various ligands. Researchers hope to show how these conformations can be utilized in potential treatments of a wide array [https://en.wikipedia.org/wiki/List_of_mental_disorders disorders]. <ref name="Drug">PMID: 24628305</ref> | Of the fifteen human class B GPCRs, eight have been confirmed as potential [https://en.wikipedia.org/wiki/Biological_target drug target]. <ref name="Drug">PMID: 24628305</ref> However, overall family B GPCRS have been difficult drug targets. This difficulty is partially related to the inherent flexibility in the class B GCGR 7TM. The flexibility comes from the ability of GCGR to be a receptor many ligands. The ECD and it's role in interactions on the extracellular side of receptors may provide evidence to how class B receptors adjust the conformational spectra for various ligands. Researchers hope to show how these conformations can be utilized in potential treatments of a wide array [https://en.wikipedia.org/wiki/List_of_mental_disorders disorders]. <ref name="Drug">PMID: 24628305</ref> | ||
=Potential Inhibitors for GCGR= | =Potential Inhibitors for GCGR= | ||
| - | There are potential class B GCGR [https://en.wikipedia.org/wiki/Enzyme_inhibitor inhibitors] that have clinic relevancy. These inhibitors to class B GCGRs have primarily focused on [https://en.wikipedia.org/wiki/Allosteric_regulation allosteric inhibitors] with high specificity and the ability to treat diseases including: [https://en.wikipedia.org/wiki/Stress-related_disorders stress disorders], managing [http://www.webmd.com/diabetes/guide/diabetes-hyperglycemia hyperglycemia], and also alternative mechanisms for treating [https://en.wikipedia.org/wiki/Migraine migraines]. <ref name="Inhibitors">PMID: 24189067</ref> Inhibitors include [https://en.wikipedia.org/wiki/Monoclonal_antibody monoclonal antibodies] which inhibit glucagon receptors through an allosteric mechanism. <ref name="Last">PMID: 19305799</ref> | + | There are potential class B GCGR [https://en.wikipedia.org/wiki/Enzyme_inhibitor inhibitors] that have clinic relevancy. These inhibitors to class B GCGRs have primarily focused on [https://en.wikipedia.org/wiki/Allosteric_regulation allosteric inhibitors] with high specificity and the ability to treat diseases including: [https://en.wikipedia.org/wiki/Stress-related_disorders stress disorders], managing [http://www.webmd.com/diabetes/guide/diabetes-hyperglycemia hyperglycemia], and also alternative mechanisms for treating [https://en.wikipedia.org/wiki/Migraine migraines]. <ref name="Inhibitors">PMID: 24189067</ref> Inhibitors include [https://en.wikipedia.org/wiki/Monoclonal_antibody monoclonal antibodies] which inhibit glucagon receptors through an allosteric mechanism. <ref name="Last">PMID: 19305799</ref> There is further research still to be done on the GCGR. |
==Research== | ==Research== | ||
Determining the structure of class B GCGRs is a reason for its lack of advanced knowledge in the field, but [https://en.wikipedia.org/wiki/X-ray_crystallography X-ray crystallography] and [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance NMR] have been the main processes performed and have had some success with it over the past couple years. <ref name="Lastt">PMID: 26227798</ref> X-ray crystallography displayed the [https://en.wikipedia.org/wiki/Crystal_structure crystal structure] of ECDs of class B GPCRs in complex with their [https://en.wikipedia.org/wiki/Ligand ligands] along with the crystal structure of the 7TM. In addition to this, NMR has allowed the ability to directly understand structures of soluble amino-terminal domains of numerous members of the secretin-like family that bind [https://en.wikipedia.org/wiki/Peptide_hormone peptide hormones]. Primary sequences analysis have led to the finding of seven segments of eighteen or more relatively hydrophobic residues that are believed to represent transmembrane helices that take part in creating an intramembranous helical bundle. <ref name="Lastt">PMID: 26227798</ref> Also, [https://en.wikipedia.org/wiki/Mutagenesis mutagenesis] has been used to determine which residues were necessary in maximizing affinity for glucagon. Finally, the orientation and mechanism of the peptide interactions within these structures are studied using peptide structure-activity relationships (SAR), receptor and ligand fragments, chimeric receptors, site-directed mutagenesis, photochemical cross-linking, and molecular modeling. <ref name="Lastt">PMID: 26227798</ref> | Determining the structure of class B GCGRs is a reason for its lack of advanced knowledge in the field, but [https://en.wikipedia.org/wiki/X-ray_crystallography X-ray crystallography] and [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance NMR] have been the main processes performed and have had some success with it over the past couple years. <ref name="Lastt">PMID: 26227798</ref> X-ray crystallography displayed the [https://en.wikipedia.org/wiki/Crystal_structure crystal structure] of ECDs of class B GPCRs in complex with their [https://en.wikipedia.org/wiki/Ligand ligands] along with the crystal structure of the 7TM. In addition to this, NMR has allowed the ability to directly understand structures of soluble amino-terminal domains of numerous members of the secretin-like family that bind [https://en.wikipedia.org/wiki/Peptide_hormone peptide hormones]. Primary sequences analysis have led to the finding of seven segments of eighteen or more relatively hydrophobic residues that are believed to represent transmembrane helices that take part in creating an intramembranous helical bundle. <ref name="Lastt">PMID: 26227798</ref> Also, [https://en.wikipedia.org/wiki/Mutagenesis mutagenesis] has been used to determine which residues were necessary in maximizing affinity for glucagon. Finally, the orientation and mechanism of the peptide interactions within these structures are studied using peptide structure-activity relationships (SAR), receptor and ligand fragments, chimeric receptors, site-directed mutagenesis, photochemical cross-linking, and molecular modeling. <ref name="Lastt">PMID: 26227798</ref> | ||
Revision as of 04:08, 19 April 2016
References
- ↑ 1.0 1.1 Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014 Jan;35(1):12-22. doi: 10.1016/j.tips.2013.11.001. Epub, 2013 Dec 18. PMID:24359917 doi:http://dx.doi.org/10.1016/j.tips.2013.11.001
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ 3.0 3.1 3.2 3.3 Yang L, Yang D, de Graaf C, Moeller A, West GM, Dharmarajan V, Wang C, Siu FY, Song G, Reedtz-Runge S, Pascal BD, Wu B, Potter CS, Zhou H, Griffin PR, Carragher B, Yang H, Wang MW, Stevens RC, Jiang H. Conformational states of the full-length glucagon receptor. Nat Commun. 2015 Jul 31;6:7859. doi: 10.1038/ncomms8859. PMID:26227798 doi:http://dx.doi.org/10.1038/ncomms8859
- ↑ 4.0 4.1 4.2 4.3 4.4 Miller LJ, Dong M, Harikumar KG. Ligand binding and activation of the secretin receptor, a prototypic family B G protein-coupled receptor. Br J Pharmacol. 2012 May;166(1):18-26. doi: 10.1111/j.1476-5381.2011.01463.x. PMID:21542831 doi:http://dx.doi.org/10.1111/j.1476-5381.2011.01463.x
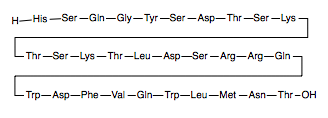
- ↑ Thomsen J, Kristiansen K, Brunfeldt K, Sundby F. The amino acid sequence of human glucagon. FEBS Lett. 1972 Apr 1;21(3):315-319. PMID:11946536
- ↑ 6.0 6.1 Bortolato A, Dore AS, Hollenstein K, Tehan BG, Mason JS, Marshall FH. Structure of Class B GPCRs: new horizons for drug discovery. Br J Pharmacol. 2014 Jul;171(13):3132-45. doi: 10.1111/bph.12689. PMID:24628305 doi:http://dx.doi.org/10.1111/bph.12689
- ↑ Mukund S, Shang Y, Clarke HJ, Madjidi A, Corn JE, Kates L, Kolumam G, Chiang V, Luis E, Murray J, Zhang Y, Hotzel I, Koth CM, Allan BB. Inhibitory mechanism of an allosteric antibody targeting the glucagon receptor. J Biol Chem. 2013 Nov 4. PMID:24189067 doi:http://dx.doi.org/10.1074/jbc.M113.496984
- ↑ Hoare SR. Allosteric modulators of class B G-protein-coupled receptors. Curr Neuropharmacol. 2007 Sep;5(3):168-79. doi: 10.2174/157015907781695928. PMID:19305799 doi:http://dx.doi.org/10.2174/157015907781695928