We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1180
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load='4L6R' size=' | + | <StructureSection load='4L6R' size='420' side='right' caption='7TM structure of human class B GPCR 4L6R', [[Resolution|resolution]] 1.80Å' scene=''> |
G protein coupled receptors (GPCRs) are recognized as the largest known class of integral membrane proteins and are divided into five families; the rhodopsin family (class A), the secretin family (class B), the adhesion family, the glutamate family (class C), and the frizzled/taste family (class F). Roughly 5% of the human genome encodes g-protein-coupled receptors which are responsible for the transduction of endogenous signals and the instigation of cellular response. The variants all contain a similar seven α-helical transmembrane domain (TMD or 7TMD) that, once bound to its peptide ligand, undergoes conformational change and tranduces a signal to coupled, heterotrimeric G proteins which initiate intracellular signal pathways and generate physiological and pathological processes. <ref>DOI 10.1371/journal.pcbi.0020013</ref> | G protein coupled receptors (GPCRs) are recognized as the largest known class of integral membrane proteins and are divided into five families; the rhodopsin family (class A), the secretin family (class B), the adhesion family, the glutamate family (class C), and the frizzled/taste family (class F). Roughly 5% of the human genome encodes g-protein-coupled receptors which are responsible for the transduction of endogenous signals and the instigation of cellular response. The variants all contain a similar seven α-helical transmembrane domain (TMD or 7TMD) that, once bound to its peptide ligand, undergoes conformational change and tranduces a signal to coupled, heterotrimeric G proteins which initiate intracellular signal pathways and generate physiological and pathological processes. <ref>DOI 10.1371/journal.pcbi.0020013</ref> | ||
| Line 16: | Line 16: | ||
Most notably, class B GPCRs contain a prominent central splay (Fig. 4) <scene name='72/727091/Corticotropin_glucagon_aligned/1'>(two Class B protein receptors demonstrating central splay)</scene> which is solvent filled and accessible from the extracellular side. This central splay is notably absent from class A GPCRs (Fig. 5) <scene name='72/727091/B2-adrenergic_glucagon_aligned/9'>(Class A vs. Class B GPCRs)</scene>, represents a tantalizing target for agonists/antagonists, and is the focus of much current research into GCGR signal regulation. <ref name= "Hollenstein 2014"/> | Most notably, class B GPCRs contain a prominent central splay (Fig. 4) <scene name='72/727091/Corticotropin_glucagon_aligned/1'>(two Class B protein receptors demonstrating central splay)</scene> which is solvent filled and accessible from the extracellular side. This central splay is notably absent from class A GPCRs (Fig. 5) <scene name='72/727091/B2-adrenergic_glucagon_aligned/9'>(Class A vs. Class B GPCRs)</scene>, represents a tantalizing target for agonists/antagonists, and is the focus of much current research into GCGR signal regulation. <ref name= "Hollenstein 2014"/> | ||
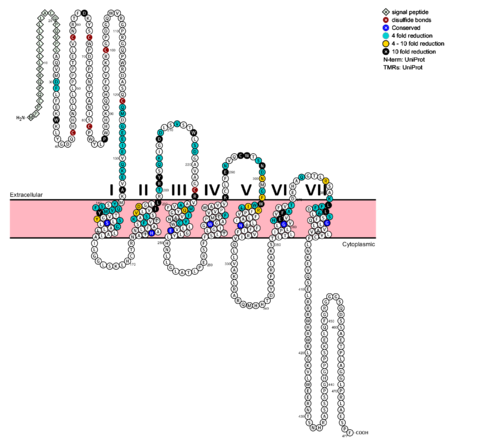
| - | [[Image:Protter GLR HUMAN.png | | + | [[Image:Protter GLR HUMAN.png |500 px|center|thumb|Fig. 6: Snake Plot of GCGR TMD<ref name= "Siu 2013"/>]] |
The snake plot (Fig. 6) shows the conservation and effects of mutagenesis in the 7TMD structure of class B human GPCR. The highly conserved amino acids imply an importance to that functioning of the individual residues and their interactions. The amino acids which have a great impact on the function of the receptor are highlighted in teal, yellow, and black, and offer evidence that the position and interaction of the amino acid is crucial for protein function. Most of the residues that play an important role in glucagon binding face the main cavity of the 7TM structure. Mutagenesis in these positions highly compromises the functioning of the glucagon binding. | The snake plot (Fig. 6) shows the conservation and effects of mutagenesis in the 7TMD structure of class B human GPCR. The highly conserved amino acids imply an importance to that functioning of the individual residues and their interactions. The amino acids which have a great impact on the function of the receptor are highlighted in teal, yellow, and black, and offer evidence that the position and interaction of the amino acid is crucial for protein function. Most of the residues that play an important role in glucagon binding face the main cavity of the 7TM structure. Mutagenesis in these positions highly compromises the functioning of the glucagon binding. | ||
| Line 45: | Line 45: | ||
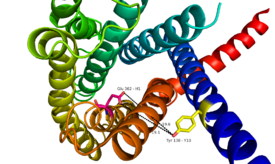
Through mutagenesis and photo-crosslinking studies, several residues deep within the central cavity of the GCGR 7TMD were discovered neighboring Glu362, which is approximately 19 angstroms from the base of the EC stalk and the location of Tyr138. (Fig. 8) | Through mutagenesis and photo-crosslinking studies, several residues deep within the central cavity of the GCGR 7TMD were discovered neighboring Glu362, which is approximately 19 angstroms from the base of the EC stalk and the location of Tyr138. (Fig. 8) | ||
| - | [[Image:Movie Frame 8.png |150 px|left|thumb|Fig.8: Relationship between Tyr138 and Glu362 - residues found to have direct relationship to glucagon binding affinity.]] | ||
Four essential residues exist deep within the central cavity which all play strong roles in ligand binding affinity. (Fig. 9) | Four essential residues exist deep within the central cavity which all play strong roles in ligand binding affinity. (Fig. 9) | ||
| Line 52: | Line 51: | ||
Essential to glucagon's binding, a long, N-terminal tail winds to a clump of 4 residues, culminating in bulge that fits into the central, anchoring site of the 7TMD. (Fig. 11) | Essential to glucagon's binding, a long, N-terminal tail winds to a clump of 4 residues, culminating in bulge that fits into the central, anchoring site of the 7TMD. (Fig. 11) | ||
| - | [[Image:Glucagon with Q3 and N-terminus.png | | + | |
| + | [[Image:Movie Frame 8.png |275 px|left|thumb|Fig.8: Relationship between Tyr138 and Glu362 - residues found to have direct relationship to glucagon binding affinity.]] | ||
| + | [[Image:Glucagon with Q3 and N-terminus.png |275 px|right|thumb|Fig. 11: Surface visualization of glucagon demonstrating three dimensional fit of N-terminal tail into binding site of GCGR central cavity active site]] | ||
==Clinical relevance== | ==Clinical relevance== | ||
Revision as of 21:03, 20 April 2016
| |||||||||||