Class B GPCRs
G protein coupled receptors (GPCRs) are the largest class of integral membrane proteins. GPCRs are divided into five families; the rhodopsin family (class A), the secretin family (class B), the glutamate family (class C), the frizzled/taste family (class F), and the adhesion family.[1] Roughly 5% of the human genome encodes g protein-coupled receptors, which are responsible for the transduction of endogenous signals and the instigation of cellular responses. All GPCRs contain a similar seven α-helical transmembrane domain that once bound to its ligand, undergoes a conformational change and tranduces a signal to coupled, heterotrimeric G proteins. The initiation of intracellular signal pathways occur in response to stimuli such as light, Ca2+, amino acids, nucleotides, odorants, peptides, and other proteins, and accomplishes many interesting physiological roles. [1]
Class B GPCRs contain 15 distinct receptors for peptide hormones and generate their signal pathway through the activation of adenylate cyclase (AC) which increases the intracellular concentration of cAMP, inositol phosphate, and calcium levels. [2] These secondary messengers are essential elements of intracellular signal cascades for human diseases including type II diabetes mellitus, osteoporosis, obesity, cancer, neurological degeneration, cardiovascular diseases, headaches, and psychiatric disorders; making their regulation through drug targeting of particular interest as disease targets. [3] Structural approaches to the development of agonists and antagonists have however been hampered by the lack of accurate Class B TMD visualizations. Recent crystal structure images of corticoptropin-releasing factor receptor 1 (PDB: 4K5Y) and human glucagon receptor (PDB: 4L6R) were accomplished through x-ray crystallography. [4] [5]
Glucagon Receptor (GCGR)
The glucagon class B GPCR (GCGR) is involved in glucose homeostasis through the binding of the signal peptide glucagon. See also PSI Structural Biology Database Glucagon is released from pancreatic α-cells when blood glucose levels fall after a period of fasting or several hours following intake of dietary carbohydrates.[6] Once the peptide hormone is released, it binds to GCGR, a 485 amino acid protein found in the liver, kidney, intestinal smooth muscle, brain, and adipose tissues. [7] Upon binding, signaling is initiated to heterotrimeric G-proteins containing Gαs. [8] GCGR can regulate additional signal pathways, including G-proteins of the Gαi family through the adoption of differing receptor conformations. [9]
Glucagon's main role is the regulation of blood glucose levels. Glucagon lowers the concentration of fructose 2,6-bisphosphate which is an allosteric inhibitor of the gluconeogenic enzyme fructose 1,6-bisphosphotase and activates phosphofructose kinase 1, which increases glucose levels via glycolysis.
Glucagon is also a regulator of the production of cholesterol, which is an energetically intensive process. When energy resources are low, downregulation of cholesterol production begins with glucagon binding to GCGR, which stimulates the phosphorylation of HMG-CoA.[6] HMG-CoA is inactivated by phosphorylation and moderates cholesterol production to conserve energy.
Glucagon also takes part in fatty acid mobilization by affecting levels of adipose tissue in the organism. Activation of GCGR by glucagon initiates triacylglycerol breakdown and the phosphorylation of perilipin and lipases via cAMP signal pathways. This allows the body to export fatty acids to the liver and other crucial tissues for energy use and makes more glucose available for use in brain functioning. [6]
Structure
The class B GPCRs, including GCGR, are different from other GPCRs in several ways. The first is that class B GPCRs contain a protrusion known as a 'stalk,' a three α-helical turn elongation of the N-terminus that protrudes past the extracellular (EC) membrane. Structural integrity of this domain in GCGR is in that A135P mutations effect stalk stability through the removal of an important salt bridge between Glu133-Lys136.
A second difference between class B and other GPCRs is that the extracellular loop 1 (ECL1) is 3-4 times longer than comparable loops in class A GPCRs, and also affects ligand binding affinity.[5]
Most notably, class B GPCRs contain a which is solvent filled and accessible from the extracellular side. This central splay is notably absent , and represents a tantalizing target for agonists/antagonists.[3]
Because of the difficulty in stabilizing and crystallizing Class B TMDs, very little is known about the conformational changes that transduce cell signals endogenously. GCGR is known to regulate additional signal pathways through the adoption of differing receptor conformations and to interact with receptor activity-modifying proteins (RAMPs) altering the signaling bias of the receptor.[9]
Glucagon Binding
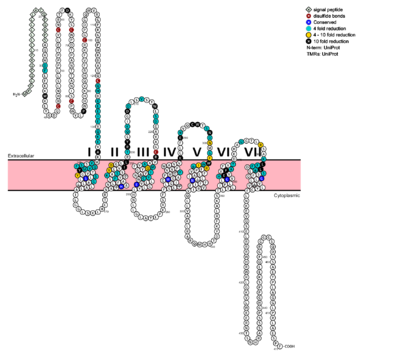
Fig. 1: Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.
[5]
In a comprehensive mutagenesis and glucagon-binding study, a total of 129 mutations were tested. 41 of these covering 28 different locations in the GCGR were found to have at least a fourfold decrease in glucagon binding affinity
[5]. (see Fig. 1) It is the face of the central cavity that harbors the majority of the residues which play an important role in glucagon binding.
[5] The binding site was shown to be a dynamic area traveling from the middle of the stalk region to deep within the 7TM core. (See Fig. 2)
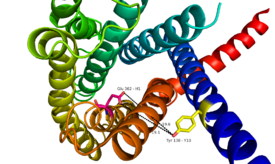
Fig. 2: Relationship between Tyr138 and Glu362 - residues spanning the height of the glucagon binding pocket in GCGR.
The binding site encompasses positions along ECL1, ECL2 and ECL3 and helices I, II, III, V, VI and VII, while extending deep in the 7TM cavity.
Glucagon itself carries essential to the proper binding of glucagon to the GCGR. Residues His 1, Gln 3, Phe 6, and Tyr 10 are critical to successful ligand
It has been discovered that the large, soluble N-terminal extracellular domains (ECD) of GCGR are primary in ligand selectivity with the deep, ligand pocket (Fig. 7) of the TMD providing secondary recognition. [7]
GPCR activity is regularly quantified by ligand binding affinity, potency, efficacy, and kinetics. These measurement are used to measure drug ligand interactions in vivo. Recently, GPCRs have been crystallized and catalogued, which tend to include a need to stabilize the receptor, emphasizing the instability of the G coupled protein receptor. Zhang et. al. imply the importance of receptor folding in the cell membrane, in the human class B GPCR the 7TM portion, for receptor stability and function. [10]
Essential, conserved residues of glucagon, as discovered through mutagenesis and photo cross-linking studies have been labeled and colored in red. [5]
Through mutagenesis and photo-crosslinking studies, several residues deep within the central cavity of the GCGR 7TMD were discovered neighboring Glu362, which is approximately 19 angstroms from the base of the EC stalk and the location of Tyr138. (Fig. 8)
Four essential residues exist deep within the central cavity which all play strong roles in ligand binding affinity. (Fig. 9)
A narrow entry gives way to a large, anchoring site for residues 1-4 of glucagon. (Fig. 10)
Essential to glucagon's binding, a long, N-terminal tail winds to a clump of 4 residues, culminating in bulge that fits into the central, anchoring site of the 7TMD. (Fig. 11)

Fig. 10: Ballooned pocket functioning as anchoring site for glucagon residues 1-4.

Fig. 11: Surface visualization of glucagon demonstrating three dimensional fit of N-terminal tail into binding site of GCGR central cavity active site
Clinical relevance
Class B secretin-like receptors have gained relevance in therapeutics and drug targets. Maintaining information about the class B GPCRs conformational flexibility, allows for a better understanding of the receptor-ligand binding and its pharmaceutical relevance. The 7TM structure offers a direct connect between the extracellular and intracellular region, which offers a mechanism for signal transduction within the cell. GPCRs regulate cellular processes as required by the organs in which they are located. GPCR’s are used in the functioning of neuron synapses, ion transport regulation, homeostasis, cell division, and cell morphology. Mutations in the GPCR have been linked with retinitis pigmentosa, female infertility, nephrogenic diabetes insipidus, and familial exudative vitreoretinopathy. [11]
A variety of small molecule modulators have been developed over the past several years providing the promise of enhanced pharmaceutical regulation of GCGR. [7](Fig's. 12 and 13)

Fig. 12: Small molecule regulators of GCGR, part 1
[7].

Fig. 13: Small molecule regulators of GCGR, part 2
[7].
Utilizing the visualizations of the GCGR 7TMD and glucagon peptide ligand, dimensional/structural analyses can be performed to develop models for novel molecules of increasing specificity for GCGR binding/regulation. Performing a dimensional analysis between the binding pocket and the base of the EC stalk, a large pseudopeptide molecule of 17-24 angstroms in size could be utilized to mimic the characteristics of GCGR's natural ligand, glucagon. (Fig's. 14 and 15)
See Also
PSI Structural Biology Database
G protein-coupled receptors Wikipedia page
References
- ↑ 1.0 1.1 Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006 Feb;2(2):e13. Epub 2006 Feb 17. PMID:16485037 doi:http://dx.doi.org/10.1371/journal.pcbi.0020013
- ↑ Bortolato A, Dore AS, Hollenstein K, Tehan BG, Mason JS, Marshall FH. Structure of Class B GPCRs: new horizons for drug discovery. Br J Pharmacol. 2014 Jul;171(13):3132-45. doi: 10.1111/bph.12689. PMID:24628305 doi:http://dx.doi.org/10.1111/bph.12689
- ↑ 3.0 3.1 Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class B GPCRs. Trends Pharmacol Sci. 2014 Jan;35(1):12-22. doi: 10.1016/j.tips.2013.11.001. Epub, 2013 Dec 18. PMID:24359917 doi:http://dx.doi.org/10.1016/j.tips.2013.11.001
- ↑ Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, Jazayeri A, Cooke RM, Weir M, Marshall FH. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013 Jul 25;499(7459):438-43. doi: 10.1038/nature12357. Epub 2013 Jul 17. PMID:23863939 doi:http://dx.doi.org/10.1038/nature12357
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013 Jul 25;499(7459):444-9. doi: 10.1038/nature12393. Epub 2013 Jul 17. PMID:23863937 doi:10.1038/nature12393
- ↑ 6.0 6.1 6.2 'Lehninger A., Nelson D.N, & Cox M.M. (2008) Lehninger Principles of Biochemistry. W. H. Freeman, fifth edition.'
- ↑ 7.0 7.1 7.2 7.3 7.4 Yang DH, Zhou CH, Liu Q, Wang MW. Landmark studies on the glucagon subfamily of GPCRs: from small molecule modulators to a crystal structure. Acta Pharmacol Sin. 2015 Sep;36(9):1033-42. doi: 10.1038/aps.2015.78. Epub 2015, Aug 17. PMID:26279155 doi:http://dx.doi.org/10.1038/aps.2015.78
- ↑ Ahren B. Islet G protein-coupled receptors as potential targets for treatment of type 2 diabetes. Nat Rev Drug Discov. 2009 May;8(5):369-85. doi: 10.1038/nrd2782. Epub 2009 Apr, 14. PMID:19365392 doi:http://dx.doi.org/10.1038/nrd2782
- ↑ 9.0 9.1 Xu Y, Xie X. Glucagon receptor mediates calcium signaling by coupling to G alpha q/11 and G alpha i/o in HEK293 cells. J Recept Signal Transduct Res. 2009 Dec;29(6):318-25. doi:, 10.3109/10799890903295150. PMID:19903011 doi:http://dx.doi.org/10.3109/10799890903295150
- ↑ Zhang X, Stevens RC, Xu F. The importance of ligands for G protein-coupled receptor stability. Trends Biochem Sci. 2015 Feb;40(2):79-87. doi: 10.1016/j.tibs.2014.12.005. Epub, 2015 Jan 15. PMID:25601764 doi:http://dx.doi.org/10.1016/j.tibs.2014.12.005
- ↑ Salon JA, Lodowski DT, Palczewski K. The significance of G protein-coupled receptor crystallography for drug discovery. Pharmacol Rev. 2011 Dec;63(4):901-37. doi: 10.1124/pr.110.003350. PMID:21969326 doi:http://dx.doi.org/10.1124/pr.110.003350






