User:R. Jeremy Johnson/Glutamate Receptor
From Proteopedia
(Difference between revisions)
(New page: <StructureSection load='4oo9' size='340' side='right' caption='metabotropic Glutamate Receptor 5 PDB:[http://www.rcsb.org/pdb/explore/explore.do?structureId=4oo9 4oo9]' scene='72/726409/Ov...) |
|||
| Line 2: | Line 2: | ||
= metabotropic Glutamate Receptor 5 = | = metabotropic Glutamate Receptor 5 = | ||
== Introduction == | == Introduction == | ||
| - | G-coupled protein receptors [https://en.wikipedia.org/wiki/G_protein–coupled_receptor (GPCR's)] are helical trans-membrane proteins that bind to an extracellular signal and activate a cellular response. The human genome encodes for approximately 750 GPCR's, 350 of which are known to respond to extracellular ligands.<ref name="GPCRRep">PMID: 12679517 </ref> GPCR's are divided into four major classes based on sequence similarity and transduction mechanism: Classes A, B, C, and F.<ref name="MSGPCR">PMID:23407534</ref> Metabotropic Glutamate Receptor 5 (<scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene>) is a class C GPCR that is involved in the G<sub>q</sub> pathway.<ref name="CCGPCR">PMID:12782243</ref> In this pathway, glutamate binds to the extracellular domain of mGlu<sub>5</sub>, and the trans-membrane domain then undergoes a conformational change that activates the coupled [http://proteopedia.org/wiki/index.php/GTP-binding_protein G-protein] on the intracellular side of the membrane.<ref name="Primary">PMID: 25042998 </ref> The activated G-protein disassociates, and the alpha subunit activates [https://en.wikipedia.org/wiki/Phospholipase_C Phospholipase C]. Phospholipase C in turn cleaves [https://en.wikipedia.org/wiki/Phosphatidylinositol_4,5-bisphosphate PIP2] to [https://en.wikipedia.org/wiki/Diglyceride DAG] and [https://en.wikipedia.org/wiki/Inositol_trisphosphate IP<sub>3</sub>]. IP<sub>3</sub> then binds to calcium channels on the [https://en.wikipedia.org/wiki/Endoplasmic_reticulum endoplasmic reticulum], creating an increased cellular concentration of calcium. Increased calcium concentrations thus lead to increased neuronal activity.<ref name="MSGPCR">PMID:23407534</ref> Due to its involvement in neuronal activity, mGlu<sub>5</sub> is highly expressed in neuronal and glial cells in the central nervous system, where glutamate serves as the major neurotransmitter. | ||
| - | |||
| - | == Structure == | ||
| - | === Overall Stucture === | ||
| - | <scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene> is seen as a [https://en.wikipedia.org/wiki/Protein_dimer homodimer] ''in vivo,'' with each subunit being comprised of three domains: extracellular, trans-membrane, and cysteine-rich. mGlu<sub>5</sub> is centered on the trans-membrane domain, comprised of seven α-helices all roughly parallel.<ref name="Primary">PMID: 25042998 </ref> Also displayed is Intracellular Loop (ICL) 1 which forms a short α-helix and interacts directly with a trans-membrane helix to stabilize mGlu<sub>5</sub>’s conformation. Additionally, ICL3 and Extracellular Loops (ECL) 1 and 3 all lack secondary structure. and ECL2 interacts with trans-membrane (TM) helices 1, 2, and 3 as well as ECL 1, again helping to stabilize the protein’s overall conformation.<ref name="Primary">PMID: 25042998 </ref> | ||
| - | ===Key Interactions=== | ||
| - | A number of intramolecular interactions within the trans-membrane domain stabilize the inactive conformation of mGlu<sub>5</sub>, as demonstrated by <scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene> being represented in the inactivate state. While in the inactive state, glutamate binding to mGlu<sub>5</sub> triggers a conformational change that leads to mGlu<sub>5</sub> being in the active state and hence initiates the aforementioned [https://en.wikipedia.org/wiki/Gq_alpha_subunit G<sub>q</sub> pathway]. The first of these interactions is an ionic interaction, termed the <scene name='72/726409/Ionic_lock2/2'>Ionic Lock</scene>, between Lysine 665 of TM3 and Glutamate 770 of TM6. Evidence for the importance of this interaction came through a kinetic study of mutant proteins where both residues were separately substituted with alanine, resulting in constitutive activity of the GPCR and its coupled pathway.<ref name="Primary">PMID: 25042998 </ref> A second critical interaction that stabilizes the inactive conformer is a <scene name='72/726409/Hydrogen_bond_614-668/2'>Hydrogen Bond </scene> between Serine 614 of ICL1 and Arginine 668 of TM3. Similarly, when Serine 614 was substituted with alanine, high levels of activity were seen in the mutant GPCR.<ref name="Primary">PMID: 25042998 </ref> A <scene name='72/726404/Scene_6/10'>Disulfide Bond </scene> between Cysteine 644 of TM3 and Cysteine 733 of <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene> is critical at anchoring ECL2 and is highly conserved across Class C GPCR’s.<ref name="Primary">PMID: 25042998 </ref> The ECL2's presence combined with the helical bundle of the trans-membrane domain creates a <scene name='72/726409/Electrogradient2/9'>Binding Cap</scene> that restricts entrance to the allosteric binding site within the seven trans-membrane α-helices. This restricted entrance has no effect on the natural ligand, glutamate, as glutamate binds to the extracellular domain, but this entrance dictates potential drug targets that act through allosteric modulation.<ref name="Primary">PMID: 25042998 </ref> | ||
| - | == Clinical Relevance == | ||
| - | ===Role in Diseases=== | ||
| - | mGlu<sub>5</sub> is located mainly post-synaptically and is in high abundance in the [https://en.wikipedia.org/wiki/Nucleus_accumbens nucleus accumbens], [https://en.wikipedia.org/wiki/Caudate_nucleus caudate nucleus], [https://en.wikipedia.org/wiki/Striatum striatum], [https://en.wikipedia.org/wiki/Hippocampus hippocampus] and [https://en.wikipedia.org/wiki/Cerebellum cerebellar cortex.]<ref name="Local">PMID: 8295733 </ref> These areas of the brain are highly involved in cognition, motivation, and emotion, which are essential neural functions. Diseases and other mental deficiencies arise from either an overactivation of the GPCR, which overactivates its coupled signaling pathway, or from underactivation of both the GPCR and its coupled pathway.<ref name="Diseases">PMID: 24237242</ref> Negative allosteric modulators (NAMs) work to decrease protein activity and are being studied as treatments for [https://en.wikipedia.org/wiki/Fragile_X_syndrome fragile X-syndrome], depression, anxiety and [https://en.wikipedia.org/wiki/Dyskinesia dyskinesia].<ref name="Primary">PMID: 25042998 </ref> Conversely, positive allosteric modulators (PAMs) work to increase protein activity and are being studied for the treatment of schizophrenia and cognitive disorders.<ref name="Diseases">PMID: 24237242</ref> | ||
| - | |||
| - | ===Interactions with a Negative Allosteric Modulator=== | ||
| - | The structure of mGlu<sub>5</sub> bound to the NAM [https://en.wikipedia.org/wiki/Mavoglurant Mavoglurant] demonstrates how protein activity is decreased through drug interactions. | ||
| - | <scene name='72/726404/Scene_7/6'>Mavoglurant</scene> binds within the allosteric binding site in the core of the seven trans-membrane α-helices, having passed through the restricted entrance formed by the <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene>. Bound Mavoglurant forms multiple interactions with the protein that further stabilize the inactive conformation. | ||
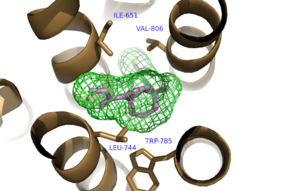
| - | The bicyclic ring system of the drug is surrounded by a pocket of mainly hydrophobic residues including Val 806, Met 802, Phe 788, Trp 785, Leu 744, Ile 651, Pro 655, and Asn 747 (Figure 1).<ref name="Primary">PMID: 25042998 </ref> The carbamate tail of Mavoglurant forms a hydrogen bond through its carbonyl oxygen to the amide side-chain of Asn 747 of TM4. A hydroxyl group similarly forms hydrogen bonds to mGlu<sub>5</sub>, specifically at two serine residues (S805 and S809) of TM7 (Figure 2). These residues form a hydrogen bonding network to other residues through their main chain atoms and a coordinated water molecule. The interactions between Mavoglurant and mGlu<sub>5</sub> involve TM helices that were not previously stabilized by any strong interactions, introducing a new level of stability that favors the inactive conformation of the protein and hence decreases the overall activity of mGlu<sub>5</sub>.<ref name="Primary">PMID: 25042998 </ref> | ||
| - | |||
| - | Mavoglurant was met by disappointing fragile X-syndrome trial results and ultimately discontinued for fragile X-syndrome in 2014, but testing Mavoglurant as a treatment for dyskinesia and [https://en.wikipedia.org/wiki/Obsessive%E2%80%93compulsive_disorder Obsessive-compulsive disorder] is still in progress. [https://en.wikipedia.org/wiki/Basimglurant Basimglurant] and [https://en.wikipedia.org/wiki/MTEP MTEP] are inhibitors that are similar to Mavoglurant as they both function as negative allosteric modulators to mGlu<sub>5</sub>, and they are currently under trials to serve as medications for depression.<ref name="Clinical">PMID: 26219727 </ref><ref name=”Clinical2”>PMID: 25043733 </ref> | ||
| - | |||
| - | [[Image:Mav_Hydrophobic_pocket.png |300 px|left|thumb|Figure 1. Hydrophobic Pocket Surrounding Mavoglurant]] | ||
| - | |||
| - | [[Image:Mav_HB_2.png|300 px|left|thumb|Figure 2. Hydrogen Bonding between mGlu<sub>5</sub> and Mavoglurant. Blue coloration represents Hydrogen Bond donor whereas red coloration represents Hydrogen Bond acceptor.]] | ||
| - | |||
| - | == Human metabotropic glutamate receptor 5 transmembrane domain == | ||
| - | StructureSection load='4oo9' size='300' frame='true' side='right' caption='Human metabotropic glutamate receptor 5 transmembrane domain bound to mavoglurant (PDB code of [http://www.rcsb.org/pdb/explore/explore.do?structureId=4oo9 4oo9]). The 7 helices comprise the bulk of the protein structure. mGlu5 receptor is an important part of the glutamate signaling pathway ' scene='72/721531/Protien_lys/3' | ||
Receiving and responding to extracellular messages is critical to the proper function of the nervous system. Glutamate is the primary excitory neurotransmitter of the Central Nervous System (CNS), and metabotropic glutamate receptor 5 is a key member of the glutamate signaling pathway<ref name="Dore" />. Metabotropic glutamate receptor 5 is a homodimeric [[GPCR]] that resides in the cellular membrane <ref name="Dore" />. mGlu5 is a member of the Class C GPCR family and can further be categorized into the Group I subgroup<ref name="Wu" />. | Receiving and responding to extracellular messages is critical to the proper function of the nervous system. Glutamate is the primary excitory neurotransmitter of the Central Nervous System (CNS), and metabotropic glutamate receptor 5 is a key member of the glutamate signaling pathway<ref name="Dore" />. Metabotropic glutamate receptor 5 is a homodimeric [[GPCR]] that resides in the cellular membrane <ref name="Dore" />. mGlu5 is a member of the Class C GPCR family and can further be categorized into the Group I subgroup<ref name="Wu" />. | ||
The functionality of the mGlu5 receptor is determined by conformational changes throughout multiple domains. mGlu5 will bind glutamate through its extracellular Venus flytrap domain and the signal will be transduced across the membrane to a heterotrimeric G protein, which will ultimately lead to calcium release and the activation of [[PKC]]<ref name="Wu" />. The signal is relayed through a Gq/11 pathway<ref name="Dore" />. Activated PKC will elicit a excitory post-synaptic response and modulate long term potentiation in the CNS<ref name="Wu" />. | The functionality of the mGlu5 receptor is determined by conformational changes throughout multiple domains. mGlu5 will bind glutamate through its extracellular Venus flytrap domain and the signal will be transduced across the membrane to a heterotrimeric G protein, which will ultimately lead to calcium release and the activation of [[PKC]]<ref name="Wu" />. The signal is relayed through a Gq/11 pathway<ref name="Dore" />. Activated PKC will elicit a excitory post-synaptic response and modulate long term potentiation in the CNS<ref name="Wu" />. | ||
| Line 31: | Line 7: | ||
Human mGlu5 is found throughout the central nervous system. Areas containing high concentrations of mGlu5 are often involved in emotional processing and higher cognition<ref name="Niswender" />. The localization of mGlu5 in the CNS and the presence of multiple domains makes mGlu5 an excellent target for treating neurological conditions including: schizophrenia, [http://www.fragilex.org/fragile-x/fragile-x-syndrome/ Fragile X], [http://www.en.wikipedia.org/wiki/Depression_(mood)depression], [http://www.en.wikipedia.org/wiki/Anxiety_disorder anxiety], and [http://www.en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease]<ref name="Wu" />. | Human mGlu5 is found throughout the central nervous system. Areas containing high concentrations of mGlu5 are often involved in emotional processing and higher cognition<ref name="Niswender" />. The localization of mGlu5 in the CNS and the presence of multiple domains makes mGlu5 an excellent target for treating neurological conditions including: schizophrenia, [http://www.fragilex.org/fragile-x/fragile-x-syndrome/ Fragile X], [http://www.en.wikipedia.org/wiki/Depression_(mood)depression], [http://www.en.wikipedia.org/wiki/Anxiety_disorder anxiety], and [http://www.en.wikipedia.org/wiki/Alzheimer's_disease Alzheimer's disease]<ref name="Wu" />. | ||
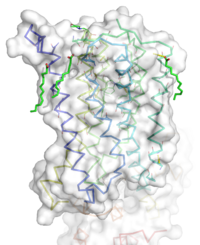
| + | [[Image:STR.png|200 px|left|thumb|'''Figure 1''': Overall Structure of the mGlu5 TMD. The polar heads on the Oleic acids orient the image with the top oriented extracellularly, the middle portion inserted into the membrane, and the lower portion oriented intracellularly. The white exterior represents the surface of the protien, and the multicolored lines interior to the surface represent the backbones 7 transmembrane alpha helices. ]] | ||
== Discovery == | == Discovery == | ||
The mGlu family of receptors was the first of the Class C [[GPCR]] to be extensively studied<ref name="Wu" />. The first regions of the protein crystallized and studied were the Venus Fly Trap domain and the cysteine -rich domain (CRD) on the extracellular region of the receptor<ref name="Dore" />.The Venus Fly Trap domain is a large extracellular domain that will selectively bind to glutamate<ref name="Wu" />. The CRD is a somewhat smaller domain composed of many ß sheets and cysteine residues <ref name="Wu" />. The CRD acts as a signal mediator between the Venus Flytrap domain and the transmembrane domain (TMD) of mGlu5, by linking to each domain with disulfide bonds<ref name="Wu" />. The hydrophobic nature and flexibility of the mGlu5 TMD made it difficult to crystallize. | The mGlu family of receptors was the first of the Class C [[GPCR]] to be extensively studied<ref name="Wu" />. The first regions of the protein crystallized and studied were the Venus Fly Trap domain and the cysteine -rich domain (CRD) on the extracellular region of the receptor<ref name="Dore" />.The Venus Fly Trap domain is a large extracellular domain that will selectively bind to glutamate<ref name="Wu" />. The CRD is a somewhat smaller domain composed of many ß sheets and cysteine residues <ref name="Wu" />. The CRD acts as a signal mediator between the Venus Flytrap domain and the transmembrane domain (TMD) of mGlu5, by linking to each domain with disulfide bonds<ref name="Wu" />. The hydrophobic nature and flexibility of the mGlu5 TMD made it difficult to crystallize. | ||
| - | Recently, the human metabotropic glutamate receptor 5 transmembrane domain was crystallized and a structure elucidated<ref name="Dore" />. Several modifications were made to the TMD for successful crystallization. The protein was thermostabilized and flexible domains were removed<ref name="Dore" />. In total residues 2-568 and 837-1153 were excised from the structure. These flexible domains allow mGlu5 to bind to its GPCR <ref name="Dore" />. The structure of the <font color='darkgreen'>'''alpha helices'''</font> are shown in <font color='darkgreen'>'''green'''</font>, and the negative allosteric modulator <span style="color:yellow;background-color:black;font-weight:bold;">mavoglurant</span> shown in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. Also in <font color='orange'><b>orange</b></font>, a T4 -<scene name='72/721531/Protien_lys/3'>Lysozyme</scene> was inserted into intracellular loop 2 to add stability<ref name="Dore" />. | + | Recently, the human metabotropic glutamate receptor 5 transmembrane domain was crystallized and a structure elucidated<ref name="Dore" />. Several modifications were made to the TMD for successful crystallization. The protein was thermostabilized and flexible domains were removed<ref name="Dore" />. In total residues 2-568 and 837-1153 were excised from the structure. These flexible domains allow mGlu5 to bind to its GPCR <ref name="Dore" />. The structure of the <font color='darkgreen'>'''alpha helices'''</font> are shown in <font color='darkgreen'>'''green'''</font>, and the negative allosteric modulator <span style="color:yellow;background-color:black;font-weight:bold;">mavoglurant</span> shown in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>. Also in <font color='orange'><b>orange</b></font>, a T4 -<scene name='72/721531/Protien_lys/3'>Lysozyme</scene> was inserted into intracellular loop 2 to add stability<ref name="Dore" />. |
| - | == Structure== | + | |
| - | + | == Structure == | |
| - | === | + | === Overall Structure === |
| - | The | + | The <scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene> TMD contains 7 <scene name='72/721531/Protien_7_helices/4'> alpha helices</scene> that spans the membrane. The protein was crystallized with <scene name='72/721531/Protien_clean_sce/6'>Oleic acid</scene> shown in <font color='red'>'''red'''</font>. On the top portion of the protein several critical extracellular loops connect the TMD to the CRD. The binding pocket can be found deep in the interior of the protein, and is mainly comprised of hydrophobic amino acids with more polar amino acids found in the upper and lower portions of the binding site<ref name="Dore" />. Inserted into the biding pocket is the negative allosteric modulator [http://www.en.wikipedia.org/wiki/Mavoglurant mavoglurant]. The TMD is in an inactive conformation, since mavoglurant is bound<ref name="Dore" />. Also, the deletion of the flexible domains leaves the mGlu5 receptor unable to bind to its [[GPCR]]<ref name="Dore" />. The inactive state is maintained by multiple ionic locks whose positions determine the active versus inactive conformation. |
| + | |||
| + | A number of intramolecular interactions within the trans-membrane domain stabilize the inactive conformation of mGlu<sub>5</sub>, as demonstrated by <scene name='72/726409/Overview/6'>mGlu<sub>5</sub></scene> being represented in the inactivate state. While in the inactive state, glutamate binding to mGlu<sub>5</sub> triggers a conformational change that leads to mGlu<sub>5</sub> being in the active state and hence initiates the aforementioned [https://en.wikipedia.org/wiki/Gq_alpha_subunit G<sub>q</sub> pathway]. The first of these interactions is an ionic interaction, termed the <scene name='72/726409/Ionic_lock2/2'>Ionic Lock</scene>, between Lysine 665 of TM3 and Glutamate 770 of TM6. Evidence for the importance of this interaction came through a kinetic study of mutant proteins where both residues were separately substituted with alanine, resulting in constitutive activity of the GPCR and its coupled pathway.<ref name="Dore"/> A second critical interaction that stabilizes the inactive conformer is a <scene name='72/726409/Hydrogen_bond_614-668/2'>Hydrogen Bond </scene> between Serine 614 of ICL1 and Arginine 668 of TM3. Similarly, when Serine 614 was substituted with alanine, high levels of activity were seen in the mutant GPCR.<ref name="Dore"/> A <scene name='72/726404/Scene_6/10'>Disulfide Bond </scene> between Cysteine 644 of TM3 and Cysteine 733 of <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene> is critical at anchoring ECL2 and is highly conserved across Class C GPCR’s.<ref name="Dore"/> The ECL2's presence combined with the helical bundle of the trans-membrane domain creates a <scene name='72/726409/Electrogradient2/9'>Binding Cap</scene> that restricts entrance to the allosteric binding site within the seven trans-membrane α-helices. This restricted entrance has no effect on the natural ligand, glutamate, as glutamate binds to the extracellular domain, but this entrance dictates potential drug targets that act through allosteric modulation.<ref name="Dore"/> | ||
| + | |||
| + | When mGlu5 is in the active conformation, signaling begins with glutamate binding to the Venus flytrap domain. The signal is transduced across the cysteine-rich domain to the TMD<ref name="Niswender" />. Next, the dimerization of the TMD occurs. This activates the Gq/11 pathway, which activates phospholipase Cβ<ref name="Niswender" />. The active [http://www.proteopedia.org/wiki/index.php/2zkm phospholipase Cβ] hydrolyzes phosphotinositides and generates [https://pubchem.ncbi.nlm.nih.gov/compound/439456#section=Top inositol 1,4,5-trisphosphate] and [http://www.sivabio.50webs.com/ip3.htm diacyl-glycerol]<ref name="Woodcock" />. This results in calcium mobilization and activation of protein kinase C ([[PKC]])<ref name="Niswender" />. Calcium is a neurotransmitter, and relatively low concentrations of calcium can cause a large response across the neuronal synapse<ref name="Niswender" />. In addition to calcium stimulating an excitory response in nerve cells, PKC can be activated for regulatory purposes by the influx of calcium. A serine on PKC can become phosphorylated, which leaves PKC unable to bind to G beta-gamma (Gßγ) protein complex<ref name="Niswender" />. Unbound Gßγ protein can then inhibit voltage-sensitive calcium channels to reduce calcium influx and provide feedback inhibition to the glutamate signaling pathway. <ref name="Niswender" />. | ||
| - | When mGlu5 is in the active conformation, signaling begins with glutamate binding to the Venus flytrap domain. The signal is transduced across the cysteine-rich domain to the TMD<ref name="Niswender" />. Next, the dimerization of the TMD occurs. This activates the Gq/11 pathway, which activates phospholipase Cβ<ref name="Niswender" />. The active [http://www.proteopedia.org/wiki/index.php/2zkm phospholipase Cβ] hydrolyzes phosphotinositides and generates [https://pubchem.ncbi.nlm.nih.gov/compound/439456#section=Top inositol 1,4,5-trisphosphate] and [http://www.sivabio.50webs.com/ip3.htm diacyl-glycerol]<ref name="Woodcock" />. This results in calcium mobilization and activation of protein kinase C ([[PKC]])<ref name="Niswender" />. Calcium is a neurotransmitter, and relatively low concentrations of calcium can cause a large response across the neuronal synapse<ref name="Niswender" />. In addition to calcium stimulating an excitory response in nerve cells, PKC can be activated for regulatory purposes by the influx of calcium. A serine on PKC can become phosphorylated, which leaves PKC unable to bind to G beta-gamma (Gßγ) protein complex<ref name="Niswender" />. Unbound Gßγ protein can then inhibit voltage-sensitive calcium channels to reduce calcium influx and provide feedback inhibition to the glutamate signaling pathway. <ref name="Niswender" />. | ||
=== Extracellular Domain === | === Extracellular Domain === | ||
The extracellular domain of mGlu5 contains several key extracellular loops that will help modulate ligand binding. <scene name='72/721532/Ecl_trail_7/2'>Extracellular Loops</scene> are shown here with extracellular loops (ECLs) <font color='purple'><b>1</b></font>, <font color='purple'><b>2</b></font>, and <font color='purple'><b>3</b></font> highlighted in <font color='purple'><b>purple</b></font>. Additionally in the ECL domain, a <scene name='72/721531/Ecl_trail_1/5'>disulfide bond</scene> is attached to both Helix 3 and the amino acid chain between Helix 5 and the N-terminus. <font color='teal'><b>Helix 3</b></font> and <font color='red'><b>Helix 5</b></font> are colored in <font color='teal'><b>teal</b></font> and <font color='red'><b>red</b></font> respectively.The <font color='blue'><b>N-terminus</b></font> is represented in <font color='blue'><b>blue</b></font>. The <span style="color:yellow;background-color:black;font-weight:bold;">disulfide bond</span> is highlighted in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>, and it is conserved in all classes of mGlu5 TMD<ref name="Wu" />. The disulfide bond is critical in maintaining the position of ECL 2 <ref name="Dore" />. ECLs and the helices are also factors that dictate how mavoglurant fits in the binding pocket <ref name="Dore" />. The position of these ECLs can change the effective size of the binding pocket through loop positioning<ref name="Dore" />. | The extracellular domain of mGlu5 contains several key extracellular loops that will help modulate ligand binding. <scene name='72/721532/Ecl_trail_7/2'>Extracellular Loops</scene> are shown here with extracellular loops (ECLs) <font color='purple'><b>1</b></font>, <font color='purple'><b>2</b></font>, and <font color='purple'><b>3</b></font> highlighted in <font color='purple'><b>purple</b></font>. Additionally in the ECL domain, a <scene name='72/721531/Ecl_trail_1/5'>disulfide bond</scene> is attached to both Helix 3 and the amino acid chain between Helix 5 and the N-terminus. <font color='teal'><b>Helix 3</b></font> and <font color='red'><b>Helix 5</b></font> are colored in <font color='teal'><b>teal</b></font> and <font color='red'><b>red</b></font> respectively.The <font color='blue'><b>N-terminus</b></font> is represented in <font color='blue'><b>blue</b></font>. The <span style="color:yellow;background-color:black;font-weight:bold;">disulfide bond</span> is highlighted in <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span>, and it is conserved in all classes of mGlu5 TMD<ref name="Wu" />. The disulfide bond is critical in maintaining the position of ECL 2 <ref name="Dore" />. ECLs and the helices are also factors that dictate how mavoglurant fits in the binding pocket <ref name="Dore" />. The position of these ECLs can change the effective size of the binding pocket through loop positioning<ref name="Dore" />. | ||
| Line 59: | Line 39: | ||
=== Ionic Locks === | === Ionic Locks === | ||
Another important structural feature is the series of <scene name='72/721531/Ionic_lock/5'>2 ionic locks</scene> on the intracellular side of the protein. Interactions between these five amino acids will form a salt bridge, which will stabilize the inactive conformation<ref name="Dore" />. The primary ionic lock forms between Glu770, Lys665, and Ser613<ref name="Dore" />. A secondary ionic lock occurs between Ser614 and Arg668<ref name="Dore" />. The purpose of these ionic locks is analogous to the ionic interactions that stabilize the T state in [[Hemoglobin]]. In the case of the TMD of mGlu5, the ionic lock is formed when the NAM mavoglurant is bound. These <scene name='72/721531/Ionic_lock/5'>ionic locks</scene> stabilize the inactive state, where the intracellular loops are stabilized facing inwards<ref name="Wu" />. This conformational change will effectively block the crevice that is involved in binding the G-protein<ref name="Wu" />. Models have suggested that, even in a glutamate bound state, the mavoglurant bound receptor would be dimerized but incapable of signaling<ref name="Wu" />. This signaling incapable mGlu5 dimer will help maintain the readiness of the pathway, while still decreasing signal response. | Another important structural feature is the series of <scene name='72/721531/Ionic_lock/5'>2 ionic locks</scene> on the intracellular side of the protein. Interactions between these five amino acids will form a salt bridge, which will stabilize the inactive conformation<ref name="Dore" />. The primary ionic lock forms between Glu770, Lys665, and Ser613<ref name="Dore" />. A secondary ionic lock occurs between Ser614 and Arg668<ref name="Dore" />. The purpose of these ionic locks is analogous to the ionic interactions that stabilize the T state in [[Hemoglobin]]. In the case of the TMD of mGlu5, the ionic lock is formed when the NAM mavoglurant is bound. These <scene name='72/721531/Ionic_lock/5'>ionic locks</scene> stabilize the inactive state, where the intracellular loops are stabilized facing inwards<ref name="Wu" />. This conformational change will effectively block the crevice that is involved in binding the G-protein<ref name="Wu" />. Models have suggested that, even in a glutamate bound state, the mavoglurant bound receptor would be dimerized but incapable of signaling<ref name="Wu" />. This signaling incapable mGlu5 dimer will help maintain the readiness of the pathway, while still decreasing signal response. | ||
| - | + | ||
| - | == | + | == Clinical Relevance == |
| + | ===Role in Diseases=== | ||
| + | mGlu<sub>5</sub> is located mainly post-synaptically and is in high abundance in the [https://en.wikipedia.org/wiki/Nucleus_accumbens nucleus accumbens], [https://en.wikipedia.org/wiki/Caudate_nucleus caudate nucleus], [https://en.wikipedia.org/wiki/Striatum striatum], [https://en.wikipedia.org/wiki/Hippocampus hippocampus] and [https://en.wikipedia.org/wiki/Cerebellum cerebellar cortex.]<ref name="Local">PMID: 8295733 </ref> These areas of the brain are highly involved in cognition, motivation, and emotion, which are essential neural functions. Diseases and other mental deficiencies arise from either an overactivation of the GPCR, which overactivates its coupled signaling pathway, or from underactivation of both the GPCR and its coupled pathway.<ref name="Diseases">PMID: 24237242</ref> Negative allosteric modulators (NAMs) work to decrease protein activity and are being studied as treatments for [https://en.wikipedia.org/wiki/Fragile_X_syndrome fragile X-syndrome], depression, anxiety and [https://en.wikipedia.org/wiki/Dyskinesia dyskinesia].<ref name="Dore"/> Conversely, positive allosteric modulators (PAMs) work to increase protein activity and are being studied for the treatment of schizophrenia and cognitive disorders.<ref name="Diseases">PMID: 24237242</ref> | ||
| + | [[Image:Mav_Hydrophobic_pocket.png |300 px|left|thumb|Figure 3. Hydrophobic Pocket Surrounding Mavoglurant]] | ||
| + | ===Interactions with a Negative Allosteric Modulator=== | ||
| + | The structure of mGlu<sub>5</sub> bound to the NAM [https://en.wikipedia.org/wiki/Mavoglurant Mavoglurant] demonstrates how protein activity is decreased through drug interactions. | ||
| + | <scene name='72/726404/Scene_7/6'>Mavoglurant</scene> binds within the allosteric binding site in the core of the seven trans-membrane α-helices, having passed through the restricted entrance formed by the <scene name='72/726409/Mavoglurant_overview2/5'>ECL2</scene>. Bound Mavoglurant forms multiple interactions with the protein that further stabilize the inactive conformation. The bicyclic ring system of the drug is surrounded by a pocket of mainly hydrophobic residues including Val 806, Met 802, Phe 788, Trp 785, Leu 744, Ile 651, Pro 655, and Asn 747 (Figure 3).<ref name="Dore"/> The carbamate tail of Mavoglurant forms a hydrogen bond through its carbonyl oxygen to the amide side-chain of Asn 747 of TM4. A hydroxyl group similarly forms hydrogen bonds to mGlu<sub>5</sub>, specifically at two serine residues (S805 and S809) of TM7. These residues form a hydrogen bonding network to other residues through their main chain atoms and a coordinated water molecule. The interactions between Mavoglurant and mGlu<sub>5</sub> involve TM helices that were not previously stabilized by any strong interactions, introducing a new level of stability that favors the inactive conformation of the protein and hence decreases the overall activity of mGlu<sub>5</sub>.<ref name="Dore"/> | ||
| + | |||
=== Fragile X === | === Fragile X === | ||
| - | Fragile X syndrome is the most common genetic cause of mental disability, and is a member of the [http://www.nimh.nih.gov/health/topics/autism-spectrum-disorders-asd/index.shtml Autism spectrum disorder] family<ref name="Bailey" />. The severity of intellectual disability can vary from patient to patient, but symptoms commonly stem from a misregulation of the mGlu1 and MGlu5 pathways<ref name="Bailey" />. Misregulation of these pathways leads to over potentiation in neural cells. <scene name='72/721531/Mavo/2'>Mavoglurant</scene> and other allosteric regulators like [http://www.en.wikipedia.org/wiki/Fenobam fenobam] have shown promise in treating Fragile X. One positive characteristic of ligands that target the TMD of mGlu5 is they tend to be more specific, thus interacting less with nonspecific brain proteins<ref name="Feng" />. Mavoglurant down regulates glutamate signaling in an attempt to decrease potentiation. Unfortunately, recent Phase 2 clinical trials have proven mavoglurant ineffective <ref name="Bailey" />. However, modulators of mGlu5 TMD are still being investigated as treatment for Parkinson's, Alzheimer's disease, and various addictions<ref name="Niswender" />. | + | Fragile X syndrome is the most common genetic cause of mental disability, and is a member of the [http://www.nimh.nih.gov/health/topics/autism-spectrum-disorders-asd/index.shtml Autism spectrum disorder] family<ref name="Bailey" />. The severity of intellectual disability can vary from patient to patient, but symptoms commonly stem from a misregulation of the mGlu1 and MGlu5 pathways<ref name="Bailey" />. Misregulation of these pathways leads to over potentiation in neural cells. <scene name='72/721531/Mavo/2'>Mavoglurant</scene> and other allosteric regulators like [http://www.en.wikipedia.org/wiki/Fenobam fenobam] have shown promise in treating Fragile X. One positive characteristic of ligands that target the TMD of mGlu5 is they tend to be more specific, thus interacting less with nonspecific brain proteins<ref name="Feng" />. Mavoglurant down regulates glutamate signaling in an attempt to decrease potentiation. Unfortunately, recent Phase 2 clinical trials have proven mavoglurant ineffective <ref name="Bailey" />. However, modulators of mGlu5 TMD are still being investigated as treatment for Parkinson's, Alzheimer's disease, and various addictions<ref name="Niswender" />. |
| + | |||
| + | StructureSection load='4oo9' size='300' frame='true' side='right' caption='Human metabotropic glutamate receptor 5 transmembrane domain bound to mavoglurant (PDB code of [http://www.rcsb.org/pdb/explore/explore.do?structureId=4oo9 4oo9]). The 7 helices comprise the bulk of the protein structure. mGlu5 receptor is an important part of the glutamate signaling pathway ' scene='72/721531/Protien_lys/3' | ||
== References == | == References == | ||
| - | <ref name="Dore">PMID: 25042998</ref> | ||
| - | <ref name="Wu">PMID: 24603153</ref> | ||
| - | <ref name="Niswender">PMID: 20055706</ref> | ||
| - | <ref name="Feng">PMID: 25762450</ref> | ||
| - | <ref name="Bailey">PMID: 26855682</ref> | ||
| - | <ref name="Woodcock">PMID: 18940816</ref> | ||
<references/> | <references/> | ||
| + | |||
== External Resources == | == External Resources == | ||
[http://www.fraxa.org/novartis-discontinues-development-mavoglurant-afq056-fragile-x-syndrome/ Novartis Fragile X trials] | [http://www.fraxa.org/novartis-discontinues-development-mavoglurant-afq056-fragile-x-syndrome/ Novartis Fragile X trials] | ||
Revision as of 16:49, 9 June 2016
| |||||||||||