This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
P53
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
---- | ---- | ||
| - | + | '''p53''' is a tumor suppressor. In the absence of cellular stress, p53 does not exert effect on cell fate, but under stress, p53 becomes activated and causes phenotypic changes in cells like senescence and apoptosis<ref>PMID:12719720</ref>. Many human cancer cells carry mutated p53<ref>PMID:20182618</ref>. The name '''p53''' refers to its apparent molecular mass. It runs as a 53 kDa molecule on SDS-PAGE. But based on calculations from its amino acid residues, p53's mass is actually 43.7 kDa. This difference may be due to the high number of proline residues in the protein, resulting in its migrating slowly on SDS-PAGE. | |
| - | The name '''p53''' refers to its apparent molecular mass. It runs as a 53 kDa molecule on SDS-PAGE. But based on calculations from its amino acid residues, p53's mass is actually 43.7 kDa. This difference may be due to the high number of proline residues in the protein, resulting in its migrating slowly on SDS-PAGE. | + | |
Human p53 is 393 amino acids long and has seven domains: | Human p53 is 393 amino acids long and has seven domains: | ||
| Line 24: | Line 23: | ||
- C-terminal domain | - C-terminal domain | ||
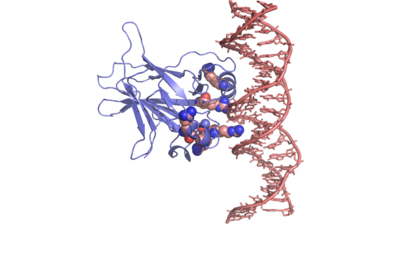
| - | p53 tumor suppressor is a | + | p53 tumor suppressor is a flexible molecule composed of four identical protein chains. Flexible molecules are difficult to study by X-ray crystallography because they do not form orderly crystals. So p53 has been studied in parts, by removing the flexible regions and solving structures of the pieces that form stable structures. The figure at the right shows the cartoon representation of the DNA-binding domain, which has been studied most. |
| - | flexible molecule composed of | + | |
| - | four identical protein chains. | + | |
| - | Flexible molecules are difficult | + | |
| - | to study by | + | |
| - | crystallography because they do | + | |
| - | not form orderly crystals. So p53 has been | + | |
| - | studied in parts, by removing | + | |
| - | the flexible regions and solving | + | |
| - | structures of the pieces that | + | |
| - | form stable structures. | + | |
| - | The figure at the right shows the cartoon representation of the DNA-binding domain, which has been studied most. | + | |
---- | ---- | ||
[[Image:P53_DNA.png | 400 px | thumb]] | [[Image:P53_DNA.png | 400 px | thumb]] | ||
| Line 123: | Line 111: | ||
* [[P53-DNA Recognition]] | * [[P53-DNA Recognition]] | ||
<br /> | <br /> | ||
| - | < | + | == References == |
| + | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 09:47, 13 June 2016
| |||||||||||
3D structures of p53 (Updated on 13-June-2016)
Additional Resources
References
- ↑ Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003 Apr;10(4):431-42. PMID:12719720 doi:http://dx.doi.org/10.1038/sj.cdd.4401183
- ↑ Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010 Feb;2(2):a001107. doi:, 10.1101/cshperspect.a001107. PMID:20182618 doi:http://dx.doi.org/10.1101/cshperspect.a001107
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Michal Harel, Eran Hodis, Mary Ball, Alexander Berchansky, David Canner