We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Reverse transcriptase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | {{STRUCTURE_3jyt| PDB=3jyt | SIZE=400| SCENE= |right|CAPTION=HIV-1 reverse transcriptase P66 subunit (grey) and P51 subunit (green) complex with DNA, GMP derivative, ATP derivative, sulfate and Mg+2 ion (green) [[1ru3]].}} | ||
| + | <StructureSection load='3jvt' size='340' side='right' caption='HIV-1 reverse transcriptase P66 subunit (grey) and P51 subunit (green) complex with DNA, GMP derivative, ATP derivative, sulfate and Mg+2 ion (green) [[1ru3]]' scene=''> | ||
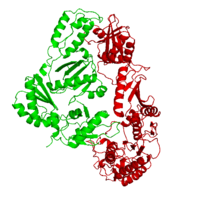
[[Image:1hmv1.png|left|200px|thumb|Crystal Structure of unliganded HIV-1 Reverse transcriptase, [[1hmv]]]] | [[Image:1hmv1.png|left|200px|thumb|Crystal Structure of unliganded HIV-1 Reverse transcriptase, [[1hmv]]]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Introduction== | ==Introduction== | ||
[[Reverse transcriptase]] (RT) or '''RNA-dependent DNA polymerase''' transcribes single-stranded RNA into double-stranded [[DNA]]. HIV-1 RT is from the human immunodeficiency virus and is a heterodimer of P66 and P51 subchains. The images at the left and at the right correspond to one representative RT structure, ''i.e.'' crystal structure of HIV-1 Reverse transcriptase ([[1hmv]]). P15 is its RNAse H domain. There are two types of inhibitors for RT: NNRTIs are the non-nucleoside inhibitors, and NRTIs are the nucleoide inhibitors. Being the protein that gives their name to Retroviruses, Reverse Transcriptase is, along with [[Hiv protease|Protease]] and Integrase, the most important part of the protein system involved in the process of infection and reproduction for viruses like HIV, MuLV and AMV. RT has the unusual property of transcribing ssRNA into dsDNA going against the Central Dogma of Molecular Biology. | [[Reverse transcriptase]] (RT) or '''RNA-dependent DNA polymerase''' transcribes single-stranded RNA into double-stranded [[DNA]]. HIV-1 RT is from the human immunodeficiency virus and is a heterodimer of P66 and P51 subchains. The images at the left and at the right correspond to one representative RT structure, ''i.e.'' crystal structure of HIV-1 Reverse transcriptase ([[1hmv]]). P15 is its RNAse H domain. There are two types of inhibitors for RT: NNRTIs are the non-nucleoside inhibitors, and NRTIs are the nucleoide inhibitors. Being the protein that gives their name to Retroviruses, Reverse Transcriptase is, along with [[Hiv protease|Protease]] and Integrase, the most important part of the protein system involved in the process of infection and reproduction for viruses like HIV, MuLV and AMV. RT has the unusual property of transcribing ssRNA into dsDNA going against the Central Dogma of Molecular Biology. | ||
| Line 53: | Line 30: | ||
One of the principal issues about this protein compared to usual DNA polymerase (besides to the similarity with the Klenow fragment), is the lack of a correction mechanism (usually made by DNA PolIII in the [[User:Karl E. Zahn/RB69 DNA polymerase (GP43)|DNA Polymerase]]); this deficiency increases the number of errors, producing more mutations and therefore giving more facultative and resistance ability to the virus. | One of the principal issues about this protein compared to usual DNA polymerase (besides to the similarity with the Klenow fragment), is the lack of a correction mechanism (usually made by DNA PolIII in the [[User:Karl E. Zahn/RB69 DNA polymerase (GP43)|DNA Polymerase]]); this deficiency increases the number of errors, producing more mutations and therefore giving more facultative and resistance ability to the virus. | ||
{{Clear}} | {{Clear}} | ||
| - | + | </StructureSection> | |
== 3D Structures of Reverse transcriptase == | == 3D Structures of Reverse transcriptase == | ||
Revision as of 09:38, 20 November 2016
| |||||||||||
3D Structures of Reverse transcriptase
Updated on 20-November-2016
See Also
- Reverse Transcriptase at Wikipedia
- Molecule of the Month (09/2002) at RCSB Protein Data Bank
- List of Reverse Transcriptase articles at Proteopedia and at RCSB Protein Data Bank
- Model of Reverse Transcriptase as one of the CBI Molecules on the Molecular Playground
- See Transcription for additional Proteopedia articles on the subject.
- For additional information, see: Human Immunodeficiency Virus
- For additional information, see: Transcription and RNA Processing
References
- ↑ Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783-90. PMID:1377403 doi:[http://dx.doi.org/10.1126/science.1377403 http://dx.doi.org/10.1126/science.1377403
- [2] Consurf Server Data Base. Evolutionary conservation profile for Reverse Transcriptase PDB file 1JLB
- [3] Abbondanzieri, E.A. et al. Nature 453, 184-189 (2008) | doi:10.1038/nature06941
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Daniel Moyano-Marino, Joel L. Sussman, Alexander Berchansky, David Canner, Amol Kapoor, Jaime Prilusky, Brian Foley, Lynmarie K Thompson, Eric Martz