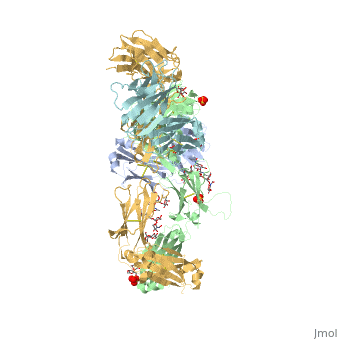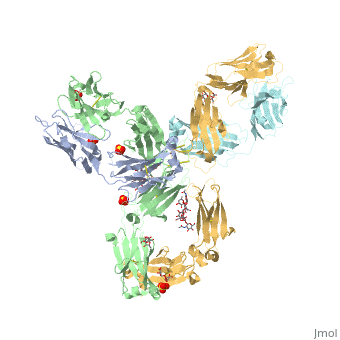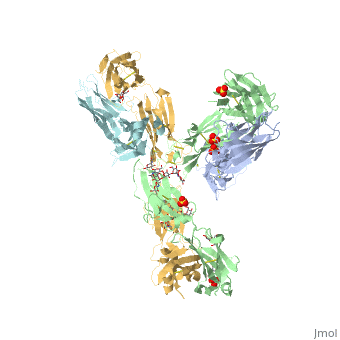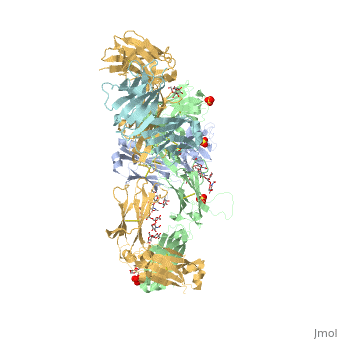We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox454
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Pembrolizumab/PD-1 Interaction == | == Pembrolizumab/PD-1 Interaction == | ||
| - | In order for pembrolizumab to block PD-1, pembrolizumab forms a large, flat paratope (antigen-binding site) that can sustain PD-1’s large epitope (where antibody attaches on antigen). The induced interaction between pembrolizumab and PD-1 gives rise to a surface conformational change on PD-1. The new structure of PD-1 becomes a very shallow, “crescent”-like shape, in contrast to it’s flat conformation when bound to PD-L1 <ref>DOI:10.1038/srep35297</ref>. | + | In order for pembrolizumab to block PD-1, pembrolizumab forms a large, flat paratope (antigen-binding site) that can sustain PD-1’s large epitope (where antibody attaches on antigen). The induced interaction between pembrolizumab and PD-1 gives rise to a surface conformational change on PD-1. The new structure of PD-1 becomes a very shallow, “crescent”-like shape, in contrast to it’s flat conformation when bound to PD-L1 <ref name="horita">DOI:10.1038/srep35297</ref>. |
== PemFv/PD-1 Interaction == | == PemFv/PD-1 Interaction == | ||
Revision as of 15:57, 15 November 2016
Pembrolizumab
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Longoria TC, Tewari KS. Evaluation of the pharmacokinetics and metabolism of pembrolizumab in the treatment of melanoma. Expert Opin Drug Metab Toxicol. 2016 Oct;12(10):1247-53. doi:, 10.1080/17425255.2016.1216976. Epub 2016 Aug 16. PMID:27485741 doi:http://dx.doi.org/10.1080/17425255.2016.1216976
- ↑ Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep. 2016 Oct 13;6:35297. doi: 10.1038/srep35297. PMID:27734966 doi:http://dx.doi.org/10.1038/srep35297
- ↑ Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep. 2016 Oct 13;6:35297. doi: 10.1038/srep35297. PMID:27734966 doi:http://dx.doi.org/10.1038/srep35297
- ↑ Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep. 2016 Oct 13;6:35297. doi: 10.1038/srep35297. PMID:27734966 doi:http://dx.doi.org/10.1038/srep35297
- ↑ Deeks ED. Pembrolizumab: A Review in Advanced Melanoma. Drugs. 2016 Mar;76(3):375-86. doi: 10.1007/s40265-016-0543-x. PMID:26846323 doi:http://dx.doi.org/10.1007/s40265-016-0543-x
- ↑ Longoria TC, Tewari KS. Evaluation of the pharmacokinetics and metabolism of pembrolizumab in the treatment of melanoma. Expert Opin Drug Metab Toxicol. 2016 Oct;12(10):1247-53. doi:, 10.1080/17425255.2016.1216976. Epub 2016 Aug 16. PMID:27485741 doi:http://dx.doi.org/10.1080/17425255.2016.1216976
- ↑ Horita S, Nomura Y, Sato Y, Shimamura T, Iwata S, Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci Rep. 2016 Oct 13;6:35297. doi: 10.1038/srep35297. PMID:27734966 doi:http://dx.doi.org/10.1038/srep35297