Transcription-repair coupling factor
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
| + | == Function == | ||
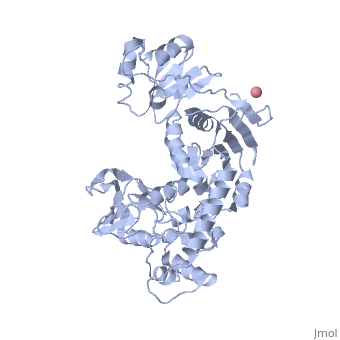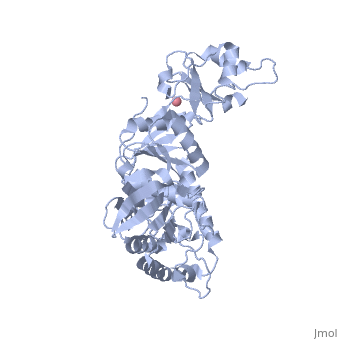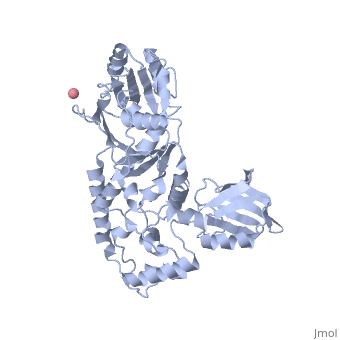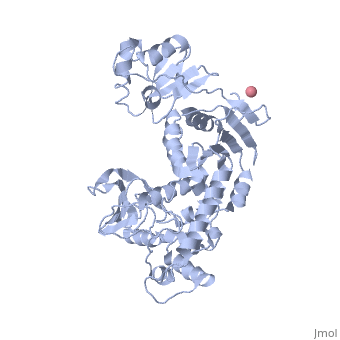
'''Transcription-repair coupling factor (TRCF)''' enables the coupling of these processes in bacteria and humans. The TRCF has ATPase activity. <scene name='2eyq/Asymmunit/2'>The crystallographic asymmetric unit of the solved structure contains 2 monomers of Mfd (residues 2-1147 of 1151 for one molecule and 5-1147 of the other), five HEPES molecules, and the sulfate ions.</scene> | '''Transcription-repair coupling factor (TRCF)''' enables the coupling of these processes in bacteria and humans. The TRCF has ATPase activity. <scene name='2eyq/Asymmunit/2'>The crystallographic asymmetric unit of the solved structure contains 2 monomers of Mfd (residues 2-1147 of 1151 for one molecule and 5-1147 of the other), five HEPES molecules, and the sulfate ions.</scene> | ||
Size exclusion chromatogaphy indicates that the protein <scene name='2eyq/Biolunit/1'>exists as a monomer in solution.</scene> | Size exclusion chromatogaphy indicates that the protein <scene name='2eyq/Biolunit/1'>exists as a monomer in solution.</scene> | ||
| - | + | == Structural highlights == | |
<scene name='2eyq/Figonea/2'>The monomer colored as</scene> an N→C rainbow similar to [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig1&_ba=1&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=a4326e4023d32f5de977da04b9a9fdf6 Figure 1A] of [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4J79KGF-J&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cd0b2f15037bec97859866e118038962 the article describing] the structure. {{Link Toggle FancyCartoonHighQualityView}}. | <scene name='2eyq/Figonea/2'>The monomer colored as</scene> an N→C rainbow similar to [http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_udi=B6WSN-4J79KGF-J&_image=fig1&_ba=1&_user=10&_rdoc=1&_fmt=full&_orig=search&_cdi=7051&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=a4326e4023d32f5de977da04b9a9fdf6 Figure 1A] of [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4J79KGF-J&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=cd0b2f15037bec97859866e118038962 the article describing] the structure. {{Link Toggle FancyCartoonHighQualityView}}. | ||
Revision as of 11:05, 27 November 2016
| |||||||||||
Reference
- Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006 Feb 10;124(3):507-20. PMID:16469698 doi:10.1016/j.cell.2005.11.045
Created with the participation of Wayne Decatur
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Michal Harel, Alexander Berchansky, Wayne Decatur