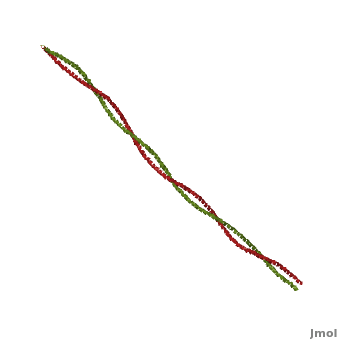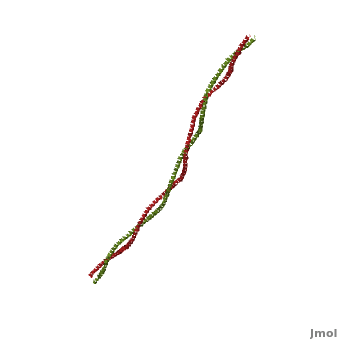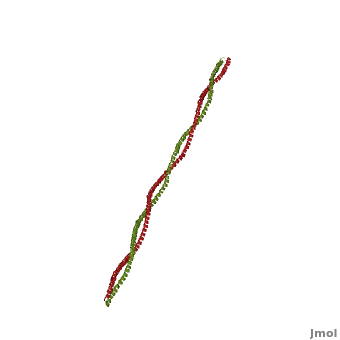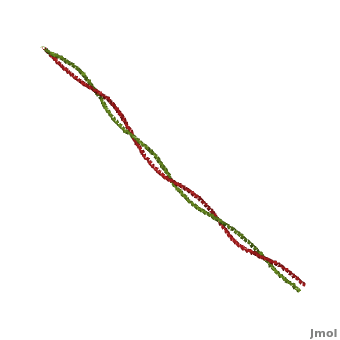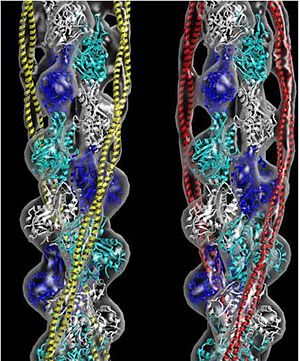
Tropomyosin (seen in yellow and red) wrapped around actin filaments, which are EM reconstructions with G-actin ribbion structures filling in the EM structure. (Picture generated from William Lehman's
Website)
Function
Tropomyosin (TM) is an actin binding protein, which consists of a coiled-coil dimer (see left) and forms a polymer along the length of actin by a head-to-tail overlap along the major grove of actin (see down & left)[1]. The head-to-tail overlap allows flexibility between the tropomyosin dimers so it will lay unstrained along the filament[1]. Each tropomyosin molecule spans seven actin monomers within a filament and lays N- to C- terminally from actin's pointed to barbed end[2]. The 284 amino acid helix has a length of 420 Angstroms and has a molecular weight around 65-70 kilodaltons (vertebrate tropomyosin)[1][3]. A few of tropomyosin's characteristics as an actin binding protein includes regulation, stabilization and recruitment.
Its role in muscle mechanics has been well established as it is a major regulatory component of the contractile apparatus, but its role in non-muscle systems is becoming evermore clear. In mammals, there are at least 40 known isoforms, which are generated by alternative splicing of multiple genes[1][2]. These isoforms in non-muscle systems contribute to actin's stability by increasing filament rigidity and protecting the filament from actin severing proteins, like gelsolin or cofilin. The different isoforms also aid in recruitment of various proteins, including myosin (a family of molecular motors)[4][5]. The reason for tropomyosin's diversity is not well known, but it is thought to exist to function at different developmental stages in some species as well as function in specific cells of higher multicellular organisms[1].
The details of its structure/function relationship defines its role in different cell systems and disease states, this page further discuss tropomyosin's structure and function. See also Fibrous Proteins.
Tropomyosin's Structure
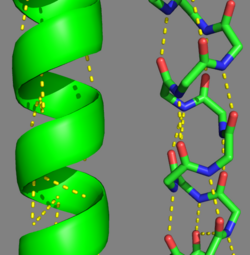
Tropomyosin's Alpha Helices are represented in ribbon (Chain A) and ball and stick form (Chain B, without side chains). The yellow lines represent the polar contacts within the protein backbone, which stabilize the helical structure.
As determined by the Structural Classification of Protein's (SCOP) Database, Tropomyosin is categorized as follows (general to specific):
- Class: coiled-coil
- Fold: parallel coiled-coil
- Superfamily: tropomyosin
- Family: pig [1c1g]
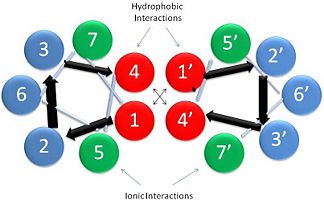
Helical Wheel Diagram Representation of Tropopomyosin: The coiled-coil dimer is stabilized by hydrophobic and ionic interactions (red=hydrophobic, blue=polar & green=charged)Image Reconstructed from
[1].
Categorization of tropomyosin's describes a unique pattern of amino acids within the primary structure of the alpha helices that comprise the dimer interface[1]. The unique amino acid pattern, found within all coiled-coil proteins, is a heptad repeat, which follows a similar pattern to: H-P-P-H-C-P-C, where H is hydrophobic, P is polar and C is charged[1][3]. This heptad repeat forms a right-handed alpha helical secondary structure (see right for alpha helix secondary structure)[1]. This alpha helix is special in that it forms a hydrophobic strip along one side, which will interface with an adjacent alpha helix that also contains the heptad repeat and hydrophobic strip. These strips aid in the dimerization of tropomyosin and is important in the characteristic coiled-coil domain.
This coiled-coil domain is represented as a helical wheel diagram (see right), whereby the four amino acids (two from each alpha chain) that are adjacent to each other contribute to a , represented in red, while the four amino acids (two from each alpha chain) flanking the hydrophobic core provide an ionic interaction, or salt-bridge represented as green. The can be observed by observing chain A, residue 26, which is a glutamic acid and forms a salt bridge with an arginine, 305 on Chain B. This weak stabilizing interaction has both and electrostatic interaction, via the ions, and two hydrogen bonds between the residues. This extra stability aids in its left-handed super-coil conformation. The two hydrogen bonds between the residues have an average distance of 2.85Å. Combined the hydrophobic and ionic interactions contribute significantly to the stability of the dimer[1][3].
The amino acids from each alpha chain interact with each other uniquely in that side chains of the helices interfacing will pack together in a knobs in holes arrangement (i + i+4 ridges fit into i + i+4 grooves)[1]. This is accomplished by the ridges of one helix fitting between the grooves (between the hydrophobic amino acids in position a & d, see helical wheel diagram) of the adjacent helix. This packing further allows for the two alpha helices to wrap tightly around each other in a left handed supercoil conformation further stabilizing the tropomyosin dimer[1].
Tropomyosin, as mentioned above, will form a long polymer along the length of actin in a head-to-tail overlap[2]. This occurs as the amino acids from the of one dimer overlaps with the amino acids of the of another dimer. There are several in the overlap region, which consist of ionic, hydrophobic and non-polar interactions[2]. (Note: many of the methionine residues interacting between the dimers are selenomethionine, which are used here to help solve the x-ray crystal structure.) Interestingly, most of the variation seen among tropomyosin isoforms is in the overlap region, which will affect polymer formation along the actin filament[2]. The actin binding sites on tropomyosin (protein-protein interactions) are currently not easily recognizable, but it is thought that a periodic repeat of seven consensus residues contribute to tropomyosin binding to actin[1]. The seven fold repeat is a mix of charged and non-polar residues in the 2,3 and 6 positions (see helical wheel diagram above)[1].
Post-Translational Modifications
There are two types of post-translational modifications to tropomyosin: phosphorylation acetylation[1]. Phosphorylation occurs on amino acid [1]. This phosphorylation is occurs as a result of oxidative stress, which is associated with actin remodeling and recruitment of additional tropomyosin into stress fibers[1]. The acetylation occurs on the N-terminus of the N-terminal methionine, which is essential for: coiled-coil stability, overlap formation and actin binding[2][1].
Evolutionary Conservation
Tropomyosin is highly conserved actin binding protein, which is found in Eukarya from the animal kingdom to yeast, with the exception of plants[1]. The earliest characterization of tropomyosin gene lineage was in yeast (budding and fission yeast)[1][6]. These genes are known as TPM1 and TPM2, respectively, and share 64.5% sequence identity[1][6]. As we move away from unicellular organisms and into multicellular invertebrates, tropomyosin genes in nematodes slightly diverge from yeast, but have 85-90% sequence identity between their genes[1]. Further analysis for vertebrates show there are four genes that generate over 40 known mammalian isoforms of tropomyosin, which are synthesized by exon splicing[1]. The slight evolution change of tropomyosin has occurred as a result of the increasing need of tropomyosin to function in different systems, but tropomyosin has evolutionarily stayed well conserved because of the basic structural pressures imposed on the protein[1]. It is interesting to note, the region with the least conservation has been in the N and C terminus. This is the result of head-tail interactions changing to accommodate different polymer confirmations along actin for different functions[1]. (To see this evolutionary conservation, go to the top right image of the web page and click on the "show" link, which is to the right of "Evolutionary Conservation".)
Tropomyosin's Function: Muscle and Non-Muscle Systems
Muscle tissue is comprised of many muscle fibers or cells. Those muscle fibers consist of myofibrils that contain a series of contractile units called sarcomeres. Within this unit contains thick filaments, comprised mainly of myosin, and thin filaments, which contain actin, tropomyosin and troponin. Tropomyosin in striated muscle systems (skeletal and cardiac) acts to inhibit the myosin cross-bridges from binding to the myosin binding site on thin filaments, this tropomyosin state is in the "Blocked" position[7]. When the muscle is stimulated, there is a rise in intracellular calcium stemming from a cascade of cellular processes. As the calcium is bathing the sarcomere, it will bind to the troponin complex, which is bound to both actin and tropomyosin[8]. The troponin will displace the tropomyosin from a "Blocked" to a "Closed" position[7]. This transition allows the myosin head to interact weakly with the myosin binding site[8]. The tropomyosin is displaced to its final position, "Open" state, along actin filament as myosin binds to its site[7]. These three tropomyosin states along the filament is referred to as the three state model[7]. As the intracellular calcium concentration falls, the troponin no longer is able to displace tropomyosin and it will transition back to the "Blocked" state[7].
In non-muscle systems, as mentioned above, tropomyosin performs a wide variety of functions inside of cells. One of the specific functions tropomyosin performs within cells is recruitment of different myosin motors. Clayton et al, 2010 showed actin decorated with tropomyosin increased myosin-V (cargo transporter) motility as opposed to bare actin[4]. It was also shown that myosin-I (cargo transporter and tension senor) lost its motility function on actin decorated with tropomyosin[4]. Further evidence from a related paper, Stark et al 2010, shows tropomyosin increases the affinity of myosin-II to actin filaments[5]. So actin decorated with tropomyosin appears to spatially regulate motor activity and thus cellular function.