We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
CRISPR-Cas
From Proteopedia
(Difference between revisions)
| Line 99: | Line 99: | ||
===CRISPR subtype I-E (Cascade)=== | ===CRISPR subtype I-E (Cascade)=== | ||
| - | *'''TthCas6e (TTHB192, Cse3)''' from ''Thermus thermophilus''. <scene name='74/742625/Cv4/21'>CRISPR endoribonuclease TthCas6e (Cse3) bound to 20 nt RNA</scene> ([[2y8w]]). Other representatives: [[1wj9]], [[2y8y]], [[2y9h]], [[3qrp]], [[3qrq]], [[3qrr]]. | ||
| - | |||
| - | *'''EcoCas6e (CasE)''' from ''Escherichia coli''. [[4dzd]] (monomer). | ||
| - | |||
| - | * '''Whole Cascade/I-E''' from ''Escherichia coli'': [[4tvx]], [[4u7u]], [[4qyz]], [[5h9f]], [[5h9e]], [[5cd4]]; see below. | ||
| - | |||
| - | ====Crystal structure of a CRISPR RNA-guided surveillance complex, Cascade, bound to a ssDNA target<ref>PMID:25123481</ref>==== | ||
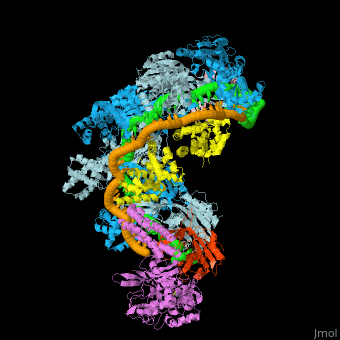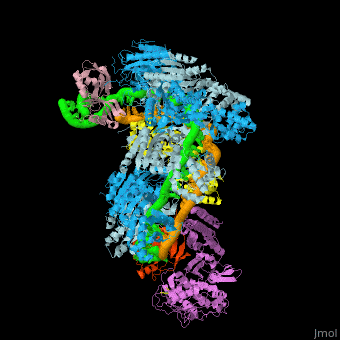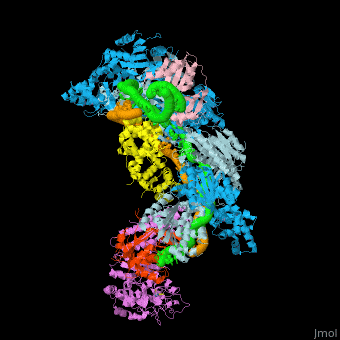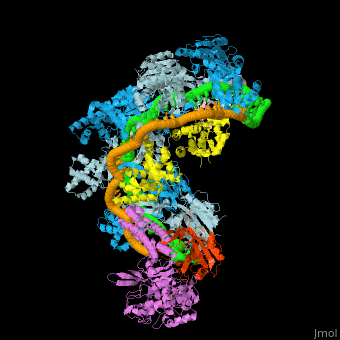
| - | The <scene name='74/742625/Cv/5'>crystal structure of ssDNA-bound Cascade has the seahorse architecture</scene>. The body is formed by a helical filament of six Cas7 subunits (Cas7.1 to 7.6) wrapped around the crRNA guide, with a head-to-tail dimer of Cse2 (Cse2.1 and Cse2.2) at the belly. Cas6e and the 3′ handle of crRNA cap the Cas7 filament at the head while Cas5 and the 5′ handle cap the tail. The N-terminal base of Cse1 is positioned at the tail of the filament; the C-terminal four-helix bundle contacts Cse2.2. The ssDNA target is juxtaposed to the guide region of the crRNA in a groove formed by the Cas7 filament, the four-helix bundle of Cse1, and the Cse2 dimer. | ||
| - | *<scene name='74/742625/Cv/6'>90° rotation about axis Z</scene>. | ||
| - | *<scene name='74/742625/Cv/7'>90° rotation about axis Y</scene>. | ||
| - | |||
| - | The <scene name='74/742625/Cv/10'>two strands of the guide-target hybrid</scene> do not twist around one another in a helix, but instead adopt an underwound ribbon-like structure reminiscent of a ladder. The 5′ and 3′ ends of the curved target strand are ~102Å apart, roughly the length of straight B-form dsDNA with an identical sequence (~107 Å). The crRNA (green) and ssDNA target (orange) are displayed in a spheres representation. Underwinding is facilitated by <scene name='74/742625/Cv/12'>kinks that occur every sixth base pair in the backbone of both strands of the hybrid</scene> (ribbon representation of the crRNA and ssDNA). <scene name='74/742625/Cv/14'>At each kink, complementary nucleotides are rotated ~90°, in opposing directions, from the axis of the duplex</scene>. Disrupted RNA and DNA nucleotides are colored red and blue, respectively. | ||
| - | |||
| - | =====Structure of the Cas7 subunit.===== | ||
| - | |||
| - | Within Cascade, the <scene name='74/742625/Cv/25'>six Cas7 subunits form a right-handed filament</scene>, with a pitch of ~135Å, around the guide target hybrid. <scene name='74/742625/Cv/16'>Rocket representation of one Cas7 colored by domain: thumb (green), fingers (blue), and palm (purple).</scene> The filament is arranged such that the <scene name='74/742625/Cv/17'>thumb of one Cas7 subunit, composed of an extended β hairpin, extends toward the fingers of the adjacent subunit</scene>. <scene name='74/742625/Cv/18'>90° rotation about axis Z</scene>. | ||
| - | |||
| - | =====Stabilization of the guide-target hybrid by Cas7, Cse1, and Cse2.===== | ||
| - | <scene name='74/742625/Cv/36'>Each 5-bp segment of the hybrid is situated between the palm of one Cas7 subunit (e.g. Cas7.2) and the fingers of the adjacent subunit (e.g. Cas7.3)</scene>. 5-bp segment is colored red. Extensive contacts between the guide region of the crRNA and the Cas7 filament bury a large portion of the crRNA backbone, leaving the bases solvent-exposed. The absence of direct contacts between protein side chains and bases of the crRNA explains the lack of sequence specificity by Cascade for the guide sequence. <scene name='74/742625/Cv/37'>Close-up view of the bound crRNA</scene>. The DNA target has been removed for clarity. Intercalation by Met166 from Cas7 is highlighted. <scene name='74/742625/Cv/38'>Several highly conserved polar and positively charged residues (Arg20, Lys27, Ser40, Gln42, Lys45, and Lys49 - colored in magenta) from the palm of one Cas7 (e.g. Cas7.3) contact the RNA backbone</scene>, while the <scene name='74/742625/Cv/39'>fingers from the adjacent Cas7 (e.g. Cas7.2) subunit (residues 109-111, 163-169, colored in plum) contact both strands of the hybrid across the minor groove</scene>. Of note, the <scene name='74/742625/Cv/40'>Thumb (colored in olive) of one Cas7 subunit (e.g. Cas7.4) pushes through the guide-target hybrid at the 1-bp gaps</scene>. <scene name='74/742625/Cv/32'>Representation of 5 thumbs protruding guide-target hybrid at the 1-bp gaps</scene>. Each displaced RNA nucleotide adopts the syn conformation, is similarly positioned above the backbone of the downstream RNA, and is contacted by <scene name='74/742625/Cv/41'>residues from both the Cas7 palm (e.g. Cas7.3 palm; Ser43 and Arg46) and thumb (e.g. Cas7.4 thumb; Thr201 and Val203)</scene>. Overview of the <scene name='74/742625/Cv1/2'>interactions between the ssDNA target and Cse2.1, Cse2.2, and Cse1</scene>. The proteins are represented as rockets, the DNA as a surface. The positions of the disrupted DNA nucleotides (royal blue) are indicated. | ||
| - | |||
| - | =====Interactions capping the tail of Cascade===== | ||
| - | <scene name='74/742625/Cv/34'>Cas5 caps the tail of the Cas7 filament at the 5′ end of the crRNA</scene>. The structure of Cascade reveals that Cas5 is structurally related to Cas7, as it consists of a palm(residues 1 to 78 and 115 to 224) and a thumb (residues 79 to 114) domain, but lacks a fingers domain. <scene name='74/742625/Cv1/3'>Rocket representation of Cas7 and Cas5 colored by domain: thumb (green), fingers (blue), and palm (purple)</scene>. Close-up view of the <scene name='74/742625/Cv/35'>interaction between Cas5, Cas7.6, and the 5′ hook</scene>. | ||
| - | |||
| - | ====Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target at 2.45 angstrom resolution<ref>PMID:26863189</ref>==== | ||
| - | |||
| - | <scene name='74/742625/Cv2/4'>Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target</scene>. | ||
| - | *<scene name='74/742625/Cv2/9'>crRNA-dsDNA hybrid</scene>. | ||
| - | *<scene name='74/742625/Cv2/7'>crRNA-dsDNA hybrid and Cascade proteins</scene>. | ||
| - | *<scene name='74/742625/Cv2/8'>Thumbs of Cas7.2-7.6 protrude crRNA-dsDNA hybrid at the 1-bp gaps</scene>. | ||
===CRISPR subtype I-F (Cascade)=== | ===CRISPR subtype I-F (Cascade)=== | ||
Revision as of 10:47, 4 December 2016
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Didovyk A, Borek B, Tsimring L, Hasty J. Transcriptional regulation with CRISPR-Cas9: principles, advances, and applications. Curr Opin Biotechnol. 2016 Aug;40:177-84. doi: 10.1016/j.copbio.2016.06.003. Epub, 2016 Jun 23. PMID:27344519 doi:http://dx.doi.org/10.1016/j.copbio.2016.06.003
- ↑ Brophy JA, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014 May;11(5):508-20. doi: 10.1038/nmeth.2926. PMID:24781324 doi:http://dx.doi.org/10.1038/nmeth.2926
- ↑ Straubeta A, Lahaye T. Zinc fingers, TAL effectors, or Cas9-based DNA binding proteins: what's best for targeting desired genome loci? Mol Plant. 2013 Sep;6(5):1384-7. doi: 10.1093/mp/sst075. Epub 2013 May 29. PMID:23718948 doi:http://dx.doi.org/10.1093/mp/sst075
- ↑ Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014 Apr;32(4):347-55. doi: 10.1038/nbt.2842. Epub 2014 Mar 2. PMID:24584096 doi:http://dx.doi.org/10.1038/nbt.2842
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci. 2015 Jan;40(1):58-66. doi: 10.1016/j.tibs.2014.10.007. Epub, 2014 Nov 18. PMID:25468820 doi:http://dx.doi.org/10.1016/j.tibs.2014.10.007
- ↑ 6.0 6.1 Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007 Mar 23;315(5819):1709-12. PMID:17379808 doi:http://dx.doi.org/10.1126/science.1138140
- ↑ 7.00 7.01 7.02 7.03 7.04 7.05 7.06 7.07 7.08 7.09 7.10 7.11 7.12 7.13 7.14 7.15 Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016 Aug 5;353(6299):aad5147. doi: 10.1126/science.aad5147. PMID:27493190 doi:http://dx.doi.org/10.1126/science.aad5147
- ↑ Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8(4):R61. PMID:17442114 doi:http://dx.doi.org/10.1186/gb-2007-8-4-r61
- ↑ 9.0 9.1 9.2 Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008 Aug 15;321(5891):960-4. doi: 10.1126/science.1159689. PMID:18703739 doi:http://dx.doi.org/10.1126/science.1159689
- ↑ Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010 Nov 4;468(7320):67-71. doi: 10.1038/nature09523. PMID:21048762 doi:http://dx.doi.org/10.1038/nature09523
- ↑ 11.0 11.1 11.2 11.3 11.4 Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015 Nov;13(11):722-36. doi: 10.1038/nrmicro3569. Epub 2015, Sep 28. PMID:26411297 doi:http://dx.doi.org/10.1038/nrmicro3569
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 12.6 12.7 12.8 Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015 Nov 5;60(3):385-97. doi: 10.1016/j.molcel.2015.10.008. Epub 2015, Oct 22. PMID:26593719 doi:http://dx.doi.org/10.1016/j.molcel.2015.10.008
- ↑ Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015 Jun 26;348(6242):1477-81. doi: 10.1126/science.aab1452. PMID:26113724 doi:http://dx.doi.org/10.1126/science.aab1452
- ↑ 14.0 14.1 14.2 Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012 Aug 17;337(6096):816-21. doi: 10.1126/science.1225829. Epub 2012, Jun 28. PMID:22745249 doi:http://dx.doi.org/10.1126/science.1225829
- ↑ Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):E2579-86. Epub 2012 Sep 4. PMID:22949671 doi:http://dx.doi.org/10.1073/pnas.1208507109
- ↑ Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013 Feb 28;152(5):1173-83. doi: 10.1016/j.cell.2013.02.022. PMID:23452860 doi:http://dx.doi.org/10.1016/j.cell.2013.02.022
- ↑ Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013 Aug;41(15):7429-37. doi: 10.1093/nar/gkt520. Epub 2013, Jun 12. PMID:23761437 doi:http://dx.doi.org/10.1093/nar/gkt520
- ↑ Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014 Jul;32(7):677-83. doi: 10.1038/nbt.2916. Epub 2014 May 18. PMID:24837660 doi:http://dx.doi.org/10.1038/nbt.2916
- ↑ Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014 May 19;12:36. doi: 10.1186/1741-7007-12-36. PMID:24884953 doi:http://dx.doi.org/10.1186/1741-7007-12-36
- ↑ Wang R, Zheng H, Preamplume G, Shao Y, Li H. The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex. Protein Sci. 2012 Mar;21(3):405-17. doi: 10.1002/pro.2028. Epub 2012 Feb 9. PMID:22238224 doi:http://dx.doi.org/10.1002/pro.2028
- ↑ Shao Y, Li H. Recognition and Cleavage of a Nonstructured CRISPR RNA by Its Processing Endoribonuclease Cas6. Structure. 2013 Feb 27. pii: S0969-2126(13)00017-8. doi:, 10.1016/j.str.2013.01.010. PMID:23454186 doi:http://dx.doi.org/10.1016/j.str.2013.01.010
- ↑ Reeks J, Sokolowski RD, Graham S, Liu H, Naismith JH, White MF. Structure of a dimeric crenarchaeal Cas6 enzyme with an atypical active site for CRISPR RNA processing. Biochem J. 2013 Mar 25. PMID:23527601 doi:10.1042/BJ20130269
- ↑ Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2013 Oct 22. PMID:24150936 doi:http://dx.doi.org/10.1093/nar/gkt922
Categories: Topic Page | Crispr | Crispr-associated | Endonuclease | Cas9 | Cas6
![Fig. 1 Overview of the CRISPR-Cas systems. (A) Architecture of class 1 (multiprotein effector complexes) and class 2 (single-protein effector complexes) CRISPR-Cas systems. (B) CRISPR-Cas adaptive immunity is mediated by CRISPR RNAs (crRNAs) and Cas proteins, which form multicomponent CRISPR ribonucleoprotein (crRNP) complexes. The first stage is adaptation, which occurs upon entry of an invading mobile genetic element (in this case, a viral genome). Cas1 (blue) and Cas2 (yellow) proteins select and process the invading DNA, and thereafter, a protospacer (orange) is integrated as a new spacer at the leader end of the CRISPR array [repeat sequences (gray) that separate similar-sized, invader-derived spacers (multiple colors)]. During the second stage, expression, the CRISPR locus is transcribed and the pre-crRNA is processed into mature crRNA guides by Cas (e.g., Cas6) or non-Cas proteins (e.g., RNase III). During the final interference stage, the Cas-crRNA complex scans invading DNA for a complementary nucleic acid target, after which the target is degraded by a Cas nuclease. From](/wiki/images/thumb/f/f2/F1r.jpg/450px-F1r.jpg)