User:Camille Zumstein/Sandbox
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Structure Rat Calcineurin== | ==Structure Rat Calcineurin== | ||
| + | |||
<StructureSection load='4il1' size='340' side='right' caption='Rat calcineurin monomer' scene=''> | <StructureSection load='4il1' size='340' side='right' caption='Rat calcineurin monomer' scene=''> | ||
| + | |||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| Line 7: | Line 9: | ||
''' Localisation and expression''': | ''' Localisation and expression''': | ||
| - | Calcineurin is a cytoplasmic protein. The catalytic subunit calcineurin A, posses two major isoforms called calcineurin Aα and Aβ. the first is widely expressed among tissues whereas the other is mainly found in the lymphoid cells.Calcineurin Aβ is therefore involved in the mediation of the immune response <ref>http://www.jimmunol.org/content/177/4/2681.full</ref> . | + | Calcineurin is a cytoplasmic protein. The catalytic subunit calcineurin A, posses two major isoforms called calcineurin Aα and Aβ. the first is widely expressed among tissues whereas the other is mainly found in the lymphoid cells.Calcineurin Aβ is therefore involved in the mediation of the immune response <ref>http://www.jimmunol.org/content/177/4/2681.full</ref>. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Function == | == Function == | ||
| - | == | + | == General structure == |
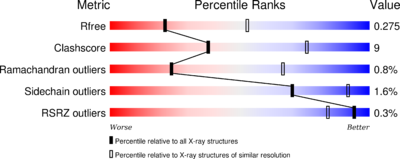
The phosphatase <scene name='75/750223/Jw_caln_sec_strcutur/1'>Calcineurin</scene> consists of 521 <ref>http://www.uniprot.org/uniprot/P63329</ref> aminoacids. The structure have been published 2013 by Qilu Ye et al. <ref>http://www.sciencedirect.com/science/article/pii/S0898656813002702</ref>. For their experiments they used [https://en.wikipedia.org/wiki/X-ray_crystallography#X-ray_diffraction x-ray diffraction]. The PDB validation obtained a Resolutionof 3.0 Å, a free R-value of 0.273 and a work R-value of 0.241 <ref>http://www.rcsb.org/pdb/explore/explore.do?structureId=4IL1</ref>. | The phosphatase <scene name='75/750223/Jw_caln_sec_strcutur/1'>Calcineurin</scene> consists of 521 <ref>http://www.uniprot.org/uniprot/P63329</ref> aminoacids. The structure have been published 2013 by Qilu Ye et al. <ref>http://www.sciencedirect.com/science/article/pii/S0898656813002702</ref>. For their experiments they used [https://en.wikipedia.org/wiki/X-ray_crystallography#X-ray_diffraction x-ray diffraction]. The PDB validation obtained a Resolutionof 3.0 Å, a free R-value of 0.273 and a work R-value of 0.241 <ref>http://www.rcsb.org/pdb/explore/explore.do?structureId=4IL1</ref>. | ||
| Line 27: | Line 24: | ||
The secondary structure includes mostly helical structures: | The secondary structure includes mostly helical structures: | ||
[[Image:170106 Uniprot SecundaryStructure.png|800 px]] | [[Image:170106 Uniprot SecundaryStructure.png|800 px]] | ||
| - | |||
== Mechanism of action == | == Mechanism of action == | ||
| + | |||
| + | |||
| + | == Addictional structural highlights == | ||
| + | |||
| + | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| + | |||
== Binding Partners == | == Binding Partners == | ||
Revision as of 10:51, 8 January 2017
Structure Rat Calcineurin
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ http://www.jimmunol.org/content/177/4/2681.full
- ↑ http://www.uniprot.org/uniprot/P63329
- ↑ http://www.sciencedirect.com/science/article/pii/S0898656813002702
- ↑ http://www.rcsb.org/pdb/explore/explore.do?structureId=4IL1
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/8811062
- ↑ http://www.uptodate.com/contents/pharmacology-of-cyclosporine-and-tacrolimus