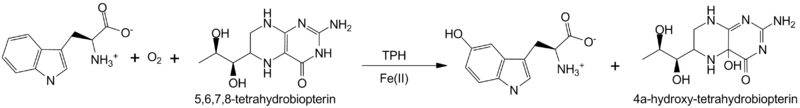We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Hydroxylase
From Proteopedia
(Difference between revisions)
| Line 44: | Line 44: | ||
**[[2v27]], [[2v28]] – PAH – ''Colwellia psychrerythraea''<br /> | **[[2v27]], [[2v28]] – PAH – ''Colwellia psychrerythraea''<br /> | ||
**[[1ltu]], [[1ltv]] – CvPAH – ''Chromobacterium violaceum''<br /> | **[[1ltu]], [[1ltv]] – CvPAH – ''Chromobacterium violaceum''<br /> | ||
| - | **[[1phz]], [[2phm]] – | + | **[[1phz]], [[2phm]] – rPAH - rat<br /> |
| + | **[[5egq]] - rPAH (mutant) <br /> | ||
**[[4bpt]] – PAH –'' Legionella pneumophila''<br /> | **[[4bpt]] – PAH –'' Legionella pneumophila''<br /> | ||
**[[1ltz]] - CvPAH + cofactor<br /> | **[[1ltz]] - CvPAH + cofactor<br /> | ||
| Line 60: | Line 61: | ||
**[[1tg2]], [[1lrm]] - hPAH (mutant) + cofactor<br /> | **[[1tg2]], [[1lrm]] - hPAH (mutant) + cofactor<br /> | ||
**[[1mmk]], [[1mmt]], [[1kw0]] - hPAH catalytic domain + cofactor + substrate<br /> | **[[1mmk]], [[1mmt]], [[1kw0]] - hPAH catalytic domain + cofactor + substrate<br /> | ||
| - | **[[4anp]] - hPAH catalytic domain + pyrimidine derivative | + | **[[4anp]] - hPAH catalytic domain + pyrimidine derivative<br /> |
| + | **[[5fii]], [[5fgi]] - hPAH residues 19-118 + phenylalanine<br /> | ||
| + | **[[5jk6]] - smPAH + Fe – slime mold<br /> | ||
| + | **[[5jk5]] - smPAH + cofactor + Fe <br /> | ||
| + | **[[5jk8]] - smPAH + cofactor + Fe + norleucine <br /> | ||
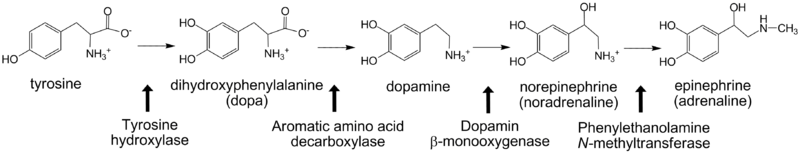
*Tyrosine hydroxylase | *Tyrosine hydroxylase | ||
| Line 69: | Line 74: | ||
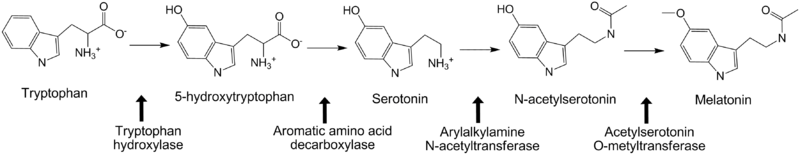
*Tryptophan hydroxylase | *Tryptophan hydroxylase | ||
| - | **[[3hf8]], [[3hfb]], [[3hf6]] – hTPH 1 catalytic domain+ inhibitor + Fe <br /> | + | **[[3hf8]], [[3hfb]], [[3hf6]], [[5tpg]], [[5l01]], [[5j6d]] – hTPH 1 catalytic domain+ inhibitor + Fe <br /> |
**[[1mlw]] – hTPH catalytic domain+ biopterin + Fe<br /> | **[[1mlw]] – hTPH catalytic domain+ biopterin + Fe<br /> | ||
**[[4v06]] - hTPH 2 catalytic domain + Fe<br /> | **[[4v06]] - hTPH 2 catalytic domain + Fe<br /> | ||
| Line 93: | Line 98: | ||
**[[1k0j]] – PaPHBH (mutant) + FAD + NADPH <br /> | **[[1k0j]] – PaPHBH (mutant) + FAD + NADPH <br /> | ||
**[[1k0l]] – PaPHBH (mutant) + FAD <br /> | **[[1k0l]] – PaPHBH (mutant) + FAD <br /> | ||
| + | **[[5tti]] – PfPHBH + FAD – ''Pseudomonas putida''<br /> | ||
| + | |||
| + | *Apartyl/asparaginyl b-hydroxylase | ||
| + | |||
| + | **[[5jz6]] – hAAH oxygenase and TPR domain + malate + Mn <br /> | ||
| + | **[[5apa]] – hAAH residues 562-758 + malate + Ni <br /> | ||
| + | **[[5jza]] – hAAH oxygenase and TPR domain + oxalylglycine + Mn <br /> | ||
| + | **[[5jz8]], [[5jzu]], [[5jzz]], [[5jqy]] – hAAH oxygenase and TPR domain + oxalylglycine + Mn + coagulation factor X peptide<br /> | ||
| + | **[[5jtc]] – hAAH oxygenase and TPR domain + pyridine dicarboxylate + Mn + coagulation factor X peptide<br /> | ||
| + | |||
| + | *Prolyl 4-hydroxylase | ||
| + | |||
| + | **[[5hv0]] - PH + Cd – ''Bacillus anthracis'' <br /> | ||
| + | **[[5c5u]] - PcvPH + Mn – Paramecium chlorella virus <br /> | ||
| + | **[[5c5t]] - PcvPH + Mn + Zn <br /> | ||
| + | **[[4p7x]] - RlPH + Co + pipecolic acid – ''Rhizobium loti'' <br /> | ||
| + | **[[4p7w]] - RlPH + Co + proline<br /> | ||
| + | |||
| + | *Salicilate hydroxylase | ||
| + | |||
| + | **[[5evy]] - PpSH + FAD + salicilate <br /> | ||
| + | |||
| + | *Dopamine b-hydroxylase | ||
| + | |||
| + | **[[4zel]] - hDH + Cu <br /> | ||
| + | |||
| + | *Ectoine hydroxylase | ||
| + | |||
| + | **[[4q5o]] - DH + Fe + 5-hydroxyectoine – ''Sphingopyxis alaskensis'' <br /> | ||
}} | }} | ||
Revision as of 08:07, 15 June 2017
| |||||||||||
3D structures of hydroxylases
Updated on 15-June-2017
Additional Resources
For additional information, see: Amino Acid Synthesis & Metabolism
References
- ↑ Fitzpatrick PF. The aromatic amino acid hydroxylases. Adv Enzymol Relat Areas Mol Biol. 2000;74:235-94. PMID:10800597
- ↑ KAUFMAN S. The enzymatic conversion of phenylalanine to tyrosine. J Biol Chem. 1957 May;226(1):511-24. PMID:13428782
- ↑ Chen D, Frey PA. Phenylalanine hydroxylase from Chromobacterium violaceum. Uncoupled oxidation of tetrahydropterin and the role of iron in hyroxylation. J Biol Chem. 1998 Oct 2;273(40):25594-601. PMID:9748224
- ↑ Leiros HK, Pey AL, Innselset M, Moe E, Leiros I, Steen IH, Martinez A. Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J Biol Chem. 2007 Jul 27;282(30):21973-86. Epub 2007 May 30. PMID:17537732 doi:10.1074/jbc.M610174200
- ↑ Grenett HE, Ledley FD, Reed LL, Woo SL. Full-length cDNA for rabbit tryptophan hydroxylase: functional domains and evolution of aromatic amino acid hydroxylases. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5530-4. PMID:3475690
- ↑ NAGATSU T, LEVITT M, UDENFRIEND S. TYROSINE HYDROXYLASE. THE INITIAL STEP IN NOREPINEPHRINE BIOSYNTHESIS. J Biol Chem. 1964 Sep;239:2910-7. PMID:14216443
- ↑ 7.0 7.1 LEVINE RJ, LOVENBERG W, SJOERDSMA A. HYDROXYLATION OF TRYPTOPHAN AND PHENYLALANINE IN NEOPLASTIC MAST CELLS OF THE MOUSE. Biochem Pharmacol. 1964 Sep;13:1283-90. PMID:14221726
- ↑ Grahame-Smith DG. Tryptophan hydroxylation in brain. Biochem Biophys Res Commun. 1964 Aug 11;16(6):586-92. PMID:5297063
- ↑ 9.0 9.1 Walther DJ, Bader M. A unique central tryptophan hydroxylase isoform. Biochem Pharmacol. 2003 Nov 1;66(9):1673-80. PMID:14563478
- ↑ Grenett HE, Ledley FD, Reed LL, Woo SL. Full-length cDNA for rabbit tryptophan hydroxylase: functional domains and evolution of aromatic amino acid hydroxylases. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5530-4. PMID:3475690
- ↑ Craig SP, Boularand S, Darmon MC, Mallet J, Craig IW. Localization of human tryptophan hydroxylase (TPH) to chromosome 11p15.3----p14 by in situ hybridization. Cytogenet Cell Genet. 1991;56(3-4):157-9. PMID:2055111
- ↑ Walther DJ, Peter JU, Bashammakh S, Hortnagl H, Voits M, Fink H, Bader M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003 Jan 3;299(5603):76. PMID:12511643 doi:http://dx.doi.org/10.1126/science.1078197
- ↑ 13.0 13.1 Patel PD, Pontrello C, Burke S. Robust and tissue-specific expression of TPH2 versus TPH1 in rat raphe and pineal gland. Biol Psychiatry. 2004 Feb 15;55(4):428-33. PMID:14960297 doi:http://dx.doi.org/10.1016/j.biopsych.2003.09.002
- ↑ Slominski A, Pisarchik A, Johansson O, Jing C, Semak I, Slugocki G, Wortsman J. Tryptophan hydroxylase expression in human skin cells. Biochim Biophys Acta. 2003 Oct 15;1639(2):80-6. PMID:14559114
- ↑ Hasegawa H, Yanagisawa M, Inoue F, Yanaihara N, Ichiyama A. Demonstration of non-neural tryptophan 5-mono-oxygenase in mouse intestinal mucosa. Biochem J. 1987 Dec 1;248(2):501-9. PMID:3435461
- ↑ Hosoda S, Nakamura W, Takatsuki K. Properties of tryptophan hydroxylase from human carcinoid tumor. Biochim Biophys Acta. 1977 May 12;482(1):27-34. PMID:16654
- ↑ Manuck SB, Flory JD, Ferrell RE, Dent KM, Mann JJ, Muldoon MF. Aggression and anger-related traits associated with a polymorphism of the tryptophan hydroxylase gene. Biol Psychiatry. 1999 Mar 1;45(5):603-14. PMID:10088047
- ↑ Mitchell JJ, Trakadis YJ, Scriver CR. Phenylalanine hydroxylase deficiency. Genet Med. 2011 Aug;13(8):697-707. doi: 10.1097/GIM.0b013e3182141b48. PMID:21555948 doi:http://dx.doi.org/10.1097/GIM.0b013e3182141b48
- ↑ Flatmark T, Stevens RC. Structural Insight into the Aromatic Amino Acid Hydroxylases and Their Disease-Related Mutant Forms. Chem Rev. 1999 Aug 11;99(8):2137-2160. PMID:11849022
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Michael Skovbo Windahl, David Canner, Joel L. Sussman, Jaime Prilusky