P53
From Proteopedia
(Difference between revisions)
| Line 33: | Line 33: | ||
In a normal cell, p53 is inactivated by its negative regulatory mdm2 (hdm2 in humans) and it is found at low levels. When DNA damage is sensed, p53's level rises. p53 binds to many regulatory sites in the genome and begins production of proteins that stop cell division until the damage is repaired. If the damage is irreparable, p53 initiates the process called programmed cell death, apoptosis, permanently removing the damage. | In a normal cell, p53 is inactivated by its negative regulatory mdm2 (hdm2 in humans) and it is found at low levels. When DNA damage is sensed, p53's level rises. p53 binds to many regulatory sites in the genome and begins production of proteins that stop cell division until the damage is repaired. If the damage is irreparable, p53 initiates the process called programmed cell death, apoptosis, permanently removing the damage. | ||
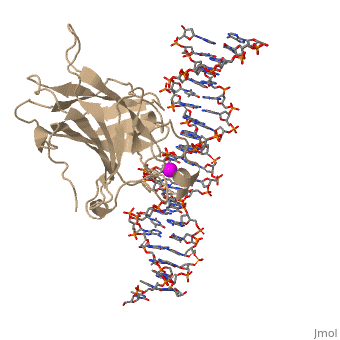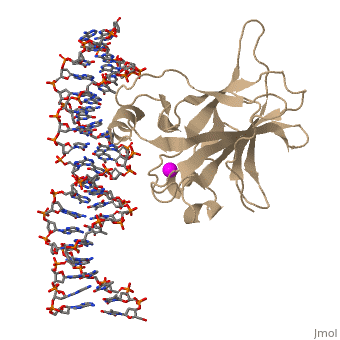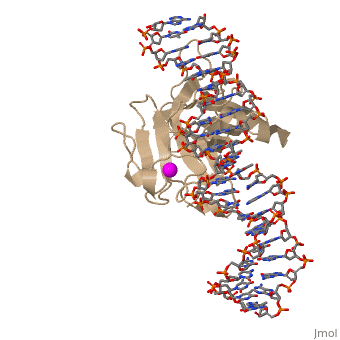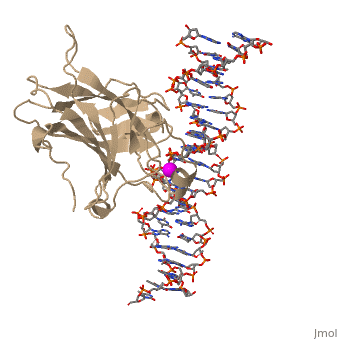
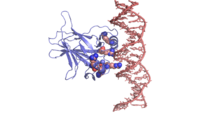
| - | In most cases of human cancer, p53 mutations have been observed. Most of the p53 mutations that may result in cancer are found in and around the DNA-binding surface of the protein. The most common mutation changes can be seen in a close up view of the <scene name='26/26327/B_chain_and_dna/5'>DNA binding domain with DNA </scene> color coded N to C (in rainbow colors), with the amino <scene name='26/26327/B_chain_and_dna/4'>R248</scene> (as space filling spheres) interacting with DNA. When mutated to another amino acid, this interaction is lost. Other key residues associated with cancer-causing mutations are <scene name='26/26327/B_chain_and_dna/8'>175, 249, 273, 282 and glycine 245</scene> | + | In most cases of human cancer, p53 mutations have been observed. Most of the p53 mutations that may result in cancer are found in and around the DNA-binding surface of the protein. The most common mutation changes can be seen in a close up view of the <scene name='26/26327/B_chain_and_dna/5'>DNA binding domain with DNA </scene> color coded N to C (in rainbow colors), with the amino <scene name='26/26327/B_chain_and_dna/4'>R248</scene> (as space filling spheres) interacting with DNA. When mutated to another amino acid, this interaction is lost. Other key residues associated with cancer-causing mutations are <scene name='26/26327/B_chain_and_dna/8'>175, 249, 273, 282 and glycine 245</scene> represented by magenta spheres. |
| Line 41: | Line 41: | ||
| - | There is a Zn-binding motif on p53. The p53 Zn atom is coordinated by residues <scene name='26/26327/B_chain_and_dna/ | + | There is a Zn-binding motif on p53. The p53 Zn atom is coordinated by residues <scene name='26/26327/B_chain_and_dna/11'>C176, H179, C238, and C242</scene> that are located on two loops, respectively. It is conceivable that the zinc plays a role in stabilizing the two loops through |
coordination. The Zn has been represented as a magenta sphere, and R248 in space filling, in the scene at the right. | coordination. The Zn has been represented as a magenta sphere, and R248 in space filling, in the scene at the right. | ||
</StructureSection> | </StructureSection> | ||
Revision as of 16:18, 14 January 2017
| |||||||||||
3D structures of p53 (Updated on 14-January-2017)
Additional Resources
References
- ↑ Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003 Apr;10(4):431-42. PMID:12719720 doi:http://dx.doi.org/10.1038/sj.cdd.4401183
- ↑ Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010 Feb;2(2):a001107. doi:, 10.1101/cshperspect.a001107. PMID:20182618 doi:http://dx.doi.org/10.1101/cshperspect.a001107
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Michal Harel, Eran Hodis, Mary Ball, Alexander Berchansky, David Canner