Dioxygenase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection size=" | + | <StructureSection size="350" scene ="Journal:JBIC:5/Opening/2" caption="Solved Crystal Structure of Hyperactive Catechol Dioxygenase"> |
'''Dioxygenases''' cleave the aromatic rings of their substrates by inserting two oxygen atoms, thus degrading these compounds. The dioxygenases are divided into 2 groups according to their mode of ring scission. The intradiol enzymes use Fe+3 as cofactor and cleave the substrate between 2 hydroxyl groups. The extradiol enzymes use Fe+2 as cofactor and cleave the substrate between a hydroxylated carbon and a non-hydroxylated one. <br /> | '''Dioxygenases''' cleave the aromatic rings of their substrates by inserting two oxygen atoms, thus degrading these compounds. The dioxygenases are divided into 2 groups according to their mode of ring scission. The intradiol enzymes use Fe+3 as cofactor and cleave the substrate between 2 hydroxyl groups. The extradiol enzymes use Fe+2 as cofactor and cleave the substrate between a hydroxylated carbon and a non-hydroxylated one. <br /> | ||
| Line 54: | Line 54: | ||
**[[1dlm]], [[1dlq]] – AcCTD + Fe+3 <br /> | **[[1dlm]], [[1dlq]] – AcCTD + Fe+3 <br /> | ||
**[[2azq]] – PpCTD + Fe+3 <br /> | **[[2azq]] – PpCTD + Fe+3 <br /> | ||
| + | **[[5umh]] – CTD – ''Burkholderia multivorans''<br /> | ||
| + | **[[5td3]] – CTD – ''Burkholderia vietnamiensis''<br /> | ||
*''CTD complex with aromatic compound'' | *''CTD complex with aromatic compound'' | ||
| Line 81: | Line 83: | ||
*Hydroxyquinol 1,2-dioxygenase (HQD) | *Hydroxyquinol 1,2-dioxygenase (HQD) | ||
| - | **[[1tmx]] – | + | **[[1tmx]] – PsiHQD + Fe+3 – ''Pimelobacter simplex''<br /> |
| + | **[[4zxd]] – PsHQD - ''Pseudomonas''<br /> | ||
| + | **[[4zxa]] – PsHQD + Cd+2 + benzonitrile <br /> | ||
| + | **[[4zxc]] – PsHQD + Fe+3 <br /> | ||
*Nitrobenzene dioxygenase (NBDO) | *Nitrobenzene dioxygenase (NBDO) | ||
| Line 93: | Line 98: | ||
**[[3t4h]], [[3t4v]], [[3t3y]] – EcAlkb + Fe+3 + inhibitor – ''Escherichia coli''<br /> | **[[3t4h]], [[3t4v]], [[3t3y]] – EcAlkb + Fe+3 + inhibitor – ''Escherichia coli''<br /> | ||
**[[2fdj]], [[2fdg]] – EcAlkb + Mn+2 + succinate <br /> | **[[2fdj]], [[2fdg]] – EcAlkb + Mn+2 + succinate <br /> | ||
| - | **[[4jht]] – EcAlkb + Mn+2 + | + | **[[4jht]] – EcAlkb + Mn+2 + quinoline derivative <br /> |
| + | **[[4rfr]] – EcAlkb + Mn+2 + rhein<br /> | ||
**[[2fdf]] – EcAlkb (mutant) + Co+2 + oxoglutarate <br /> | **[[2fdf]] – EcAlkb (mutant) + Co+2 + oxoglutarate <br /> | ||
| + | **[[4zhn]] – EcAlkb (mutant) + Co+2 + oxoglutarate + trinucleotide<br /> | ||
**[[2fd8]] – EcAlkb (mutant) + Fe+2 + oxoglutarate <br /> | **[[2fd8]] – EcAlkb (mutant) + Fe+2 + oxoglutarate <br /> | ||
**[[3khb]] – hAlkb (mutant) + Co+2 + oxoglutarate <br /> | **[[3khb]] – hAlkb (mutant) + Co+2 + oxoglutarate <br /> | ||
| Line 101: | Line 108: | ||
**[[3tht]] – hAlkb (mutant) + Mn+2 + Zn+2 + oxoglutarate <br /> | **[[3tht]] – hAlkb (mutant) + Mn+2 + Zn+2 + oxoglutarate <br /> | ||
**[[4idz]] – hFto + Ni+2 + oxalylglycine<br /> | **[[4idz]] – hFto + Ni+2 + oxalylglycine<br /> | ||
| - | **[[4ie0]], [[4ie5]] – hFto + Zn+2 + pyrimidine derivative<br /> | + | **[[4ie0]], [[4ie5]], [[4qho]] – hFto + Zn+2 + pyrimidine derivative<br /> |
**[[4ie4]], [[4ie6]] – hFto + Zn+2 + quinoline derivative<br /> | **[[4ie4]], [[4ie6]] – hFto + Zn+2 + quinoline derivative<br /> | ||
**[[4ie7]] – hFto + Zn+2 + rhein<br /> | **[[4ie7]] – hFto + Zn+2 + rhein<br /> | ||
**[[4qkn]] – hFto + Mn+2 + meclofenamic acid<br /> | **[[4qkn]] – hFto + Mn+2 + meclofenamic acid<br /> | ||
**[[4cxw]], [[4cxx]], [[4cxy]] – hFto + Ni+2 + inhibitor<br /> | **[[4cxw]], [[4cxx]], [[4cxy]] – hFto + Ni+2 + inhibitor<br /> | ||
| + | **[[5f8p]], [[5dab]] – hFto + Fe+2 + oxoglutarate + inhibitor<br /> | ||
| + | **[[4zs2]], [[4zs3]] – hFto + Mn+2 + oxoglutarate + fluorescin<br /> | ||
*''Alkb complex with DNA'' | *''Alkb complex with DNA'' | ||
| Line 141: | Line 150: | ||
**[[1f1r]], [[1f1u]] – AgHPCD + Mn+2 – ''Arthrobacter globiformis''<br /> | **[[1f1r]], [[1f1u]] – AgHPCD + Mn+2 – ''Arthrobacter globiformis''<br /> | ||
**[[1f1x]], [[1q0o]] – BfHPCD + Fe+3 – ''Brevibacterium fuscum''<br /> | **[[1f1x]], [[1q0o]] – BfHPCD + Fe+3 – ''Brevibacterium fuscum''<br /> | ||
| - | **[[2ig9]], [[3ojt]] – BfHPCD + Fe+2<br /> | + | **[[2ig9]], [[3ojt]], [[5trx]] – BfHPCD + Fe+2<br /> |
| - | **[[3ecj]], [[4ghc]] – BfHPCD (mutant) + Fe+2<br /> | + | **[[3ecj]], [[4ghc]], [[5bwg]], [[5bwh]], [[4z6l]], [[4z6m]], [[4z6n]], [[4z6o]], [[4z6p]], [[4z6q]] – BfHPCD (mutant) + Fe+2<br /> |
| + | **[[4z6z]] – BfHPCD + Fe+2 + catechol derivative<br /> | ||
| + | **[[4z6r]], [[4z6s]], [[4z6t]], [[4z6u]], [[4z6v]], [[4z6w]] – BfHPCD (mutant) + Fe+2 + catechol derivative<br /> | ||
**[[3bza]], [[3ojn]] – BfHPCD + Mn+2<br /> | **[[3bza]], [[3ojn]] – BfHPCD + Mn+2<br /> | ||
**[[3ojj]] – BfHPCD + MCo+2<br /> | **[[3ojj]] – BfHPCD + MCo+2<br /> | ||
| Line 160: | Line 171: | ||
**[[1mpy]] – PpMPC + Fe+2 <br /> | **[[1mpy]] – PpMPC + Fe+2 <br /> | ||
| - | **[[3hpv]] – PsMPC + Fe+2 | + | **[[3hpv]] – PsMPC + Fe+2 <br /> |
**[[3hpy]] – PsMPC + Fe+2 + catechol derivative<br /> | **[[3hpy]] – PsMPC + Fe+2 + catechol derivative<br /> | ||
**[[3hq0]] – PsMPC + Fe+3 + product<br /> | **[[3hq0]] – PsMPC + Fe+3 + product<br /> | ||
| Line 222: | Line 233: | ||
**[[3gzy]] – CtBDO + Fe+2 + Fe2S2 – ''Comamonas testosteroni''<br /> | **[[3gzy]] – CtBDO + Fe+2 + Fe2S2 – ''Comamonas testosteroni''<br /> | ||
**[[2xr8]] – BxBDO + Fe+2 + Fe2S2 <br /> | **[[2xr8]] – BxBDO + Fe+2 + Fe2S2 <br /> | ||
| - | **[[2xso]], [[2yfi]] – BxBDO (mutant) + Fe+2 + Fe2S2 <br /> | + | **[[2xso]], [[2yfi]], [[5aeu]] – BxBDO (mutant) + Fe+2 + Fe2S2 <br /> |
**[[1uli]] - RoBDO + Fe+2 + Fe2S2 <br /> | **[[1uli]] - RoBDO + Fe+2 + Fe2S2 <br /> | ||
**[[2gbw]] - SyBDO + Fe+2 + Fe2S2 + O2 – ''Sphingobium yanoikuyae''<br /> | **[[2gbw]] - SyBDO + Fe+2 + Fe2S2 + O2 – ''Sphingobium yanoikuyae''<br /> | ||
| Line 230: | Line 241: | ||
**[[3gzx]] – CtBDO + Fe+2 + Fe2S2 + biphenyl <br /> | **[[3gzx]] – CtBDO + Fe+2 + Fe2S2 + biphenyl <br /> | ||
**[[2xrx]] – BxBDO + Fe+2 + Fe2S2 + biphenyl <br /> | **[[2xrx]] – BxBDO + Fe+2 + Fe2S2 + biphenyl <br /> | ||
| - | **[[2xsh]] – BxBDO (mutant) + Fe+2 + Fe2S2 + biphenyl <br /> | + | **[[2xsh]], [[5aew]] – BxBDO (mutant) + Fe+2 + Fe2S2 + biphenyl <br /> |
**[[2yfj]], [[2yfl]] – BxBDO (mutant) + Fe+2 + Fe2S2 + dibenzofuran <br /> | **[[2yfj]], [[2yfl]] – BxBDO (mutant) + Fe+2 + Fe2S2 + dibenzofuran <br /> | ||
**[[1ulj]] - RoBDO + Fe+2 + Fe2S2 + biphenyl <br /> | **[[1ulj]] - RoBDO + Fe+2 + Fe2S2 + biphenyl <br /> | ||
| Line 244: | Line 255: | ||
**[[2nox]] – CmTDO + heme – ''Cupriavidus metallidurans''<br /> | **[[2nox]] – CmTDO + heme – ''Cupriavidus metallidurans''<br /> | ||
**[[4pw8]] - hTDO + Co+2 <br /> | **[[4pw8]] - hTDO + Co+2 <br /> | ||
| + | **[[5tia]], [[5ti9]] – hTDO + heme + tryptophan <br /> | ||
*Cysteine dioxygenase type I (CysDO) | *Cysteine dioxygenase type I (CysDO) | ||
**[[2gh2]], [[4ieo]], [[4iep]], [[4ieq]], [[4ier]], [[4ies]], [[4iet]], [[4ieu]], [[4iev]], [[4iew]], [[4iex]], [[4iey]], [[4iez]], [[4jtn]], [[4jto]], [[4kwj]], [[4kwk]], [[4kwl]], [[2b5h]] – rCysDO + Fe+2 – rat <br /> | **[[2gh2]], [[4ieo]], [[4iep]], [[4ieq]], [[4ier]], [[4ies]], [[4iet]], [[4ieu]], [[4iev]], [[4iew]], [[4iex]], [[4iey]], [[4iez]], [[4jtn]], [[4jto]], [[4kwj]], [[4kwk]], [[4kwl]], [[2b5h]] – rCysDO + Fe+2 – rat <br /> | ||
| - | **[[4ubg]], [[4ubh]] – rCysDO (mutant) + Fe+2 <br /> | + | **[[4ubg]], [[4ubh]], [[5ezw]], [[5efu]], [[4ysf]], [[4yyo]] – rCysDO (mutant) + Fe+2 <br /> |
| + | **[[5fdb]] – rCysDO (mutant) <br /> | ||
| + | **[[4yni]] – rCysDO <br /> | ||
| + | **[[5i0r]], [[4xet]], [[4xfb]], [[4xfc]], [[4pix]] – rCysDO (mutant) + Fe+3 <br /> | ||
**[[3eln]] – rCysDO + Fe+2 + cysteine derivative<br /> | **[[3eln]] – rCysDO + Fe+2 + cysteine derivative<br /> | ||
| + | **[[4z82]] – rCysDO (mutant) + Fe+2 + cysteine derivative<br /> | ||
| + | **[[5i0u]], [[5i0t]], [[5i0s]], [[4xf4]], [[4xf9]], [[4xfa]], [[4piz]] – rCysDO + Fe+3 + cysteine derivative<br /> | ||
| + | **[[4xf0]], [[4xf1]], [[4xfg]], [[4xfh]], [[4xfi]], [[4piy]] – rCysDO (mutant) + Fe+3 + cysteine derivative<br /> | ||
| + | **[[4xez]], [[4xff]] – rCysDO (mutant) + Fe+3 + inhibitor <br /> | ||
| + | **[[4xf3]] – rCysDO (mutant) + Fe+3 + inhibitor + cysteine<br /> | ||
**[[2q4s]], [[2atf]] – mCysDO + Ni+2 <br /> | **[[2q4s]], [[2atf]] – mCysDO + Ni+2 <br /> | ||
**[[2ic1]] – hCysDO + Fe+2 + cysteine<br /> | **[[2ic1]] – hCysDO + Fe+2 + cysteine<br /> | ||
| Line 260: | Line 280: | ||
**[[1sqd]], [[1sp9]] – AtHPPD + Fe+3 <br /> | **[[1sqd]], [[1sp9]] – AtHPPD + Fe+3 <br /> | ||
**[[1sqi]] – HPPD + Fe+3 + inhibitor - rat<br /> | **[[1sqi]] – HPPD + Fe+3 + inhibitor - rat<br /> | ||
| - | **[[1tfz]], [[1tg5]] – AtHPPD + Fe+3 + inhibitor <br /> | + | **[[1tfz]], [[1tg5]], [[5cto]] – AtHPPD + Fe+3 + inhibitor <br /> |
| - | **[[3isq]] – hHPPD + Co+2 - human<br /> | + | **[[5dhw]] – AtHPPD + Fe+3 + sulcotrione <br /> |
| + | **[[3isq]], [[5ec3]] – hHPPD + Co+2 - human<br /> | ||
**[[1sp8]] – HPPD + Fe+2 – corn<br /> | **[[1sp8]] – HPPD + Fe+2 – corn<br /> | ||
| + | **[[5hmq]] – PpHPPD + Mg+2 <br /> | ||
*Quercetin 2,3-dioxygenase (QDO) | *Quercetin 2,3-dioxygenase (QDO) | ||
| Line 281: | Line 303: | ||
**[[4j5i]] – HAD + Fe+3 – ''Mycobacterium smegmatis''<br /> | **[[4j5i]] – HAD + Fe+3 – ''Mycobacterium smegmatis''<br /> | ||
**[[3swt]] – HAD + Fe+3 – ''Mycobacterium marinum''<br /> | **[[3swt]] – HAD + Fe+3 – ''Mycobacterium marinum''<br /> | ||
| + | **[[5keu]] – BxHAD + Mg+2<br /> | ||
*3-hydroxyanthranilate 3,4-dioxygenase (HAD) | *3-hydroxyanthranilate 3,4-dioxygenase (HAD) | ||
| - | **[[1yfu]], [[4hvq]], [[4hvr]], [[4i3p]] – CmHAD + Fe+3<br /> | + | **[[4r52]] – CmHAD + Fe+2<br /> |
| + | **[[1yfu]], [[4hvq]], [[4hvr]], [[4i3p]], [[4l2n]] – CmHAD + Fe+3<br /> | ||
**[[4hvo]] – CmHAD + Fe+3 + Cu+2<br /> | **[[4hvo]] – CmHAD + Fe+3 + Cu+2<br /> | ||
**[[1yfw]] – CmHAD + Fe+3 + O2 + hydroxyanthranilate<br /> | **[[1yfw]] – CmHAD + Fe+3 + O2 + hydroxyanthranilate<br /> | ||
| Line 291: | Line 315: | ||
**[[1zvf]] – HAD + Ni+2 - yeast<br /> | **[[1zvf]] – HAD + Ni+2 - yeast<br /> | ||
**[[2qnk]] – hHAD + Ni+2 <br /> | **[[2qnk]] – hHAD + Ni+2 <br /> | ||
| + | **[[5tkq]] – hHAD + Zn+2 <br /> | ||
| + | **[[5tk5]] – hHAD + Fe+3 <br /> | ||
**[[3fe5]] – HAD + Fe+3 - bovine<br /> | **[[3fe5]] – HAD + Fe+3 - bovine<br /> | ||
| + | **[[4wzc]] – HAD + Fe+2 + enedioate derivative – ''Ralstonia metallidurans''<br /> | ||
*Carbazole 1,9a-dioxygenase (CDO) | *Carbazole 1,9a-dioxygenase (CDO) | ||
| Line 342: | Line 369: | ||
**[[4qb5]] – GBD – ''Albidiferax ferrireducens''<br /> | **[[4qb5]] – GBD – ''Albidiferax ferrireducens''<br /> | ||
**[[4rt5]] – GBD – ''Planctomyces limnophilus''<br /> | **[[4rt5]] – GBD – ''Planctomyces limnophilus''<br /> | ||
| + | |||
| + | **[[5uhj]], [[5ujp]] – GBD + Ni+2 – ''Streptomyces''<br /> | ||
| + | **[[5c4p]], [[5c68]], [[5cb9]] – TcGBD + Ni+2 – ''Thermomonospora curvata''<br /> | ||
| + | **[[5c6x]] – TcGBD + Co+2 <br /> | ||
| + | **[[4z04]] – GBD + Zn+2 + glutathione – ''Brucella abortu''s <br /> | ||
| + | |||
| + | * Procollagen-proline dioxygenase (PPD) | ||
| + | |||
| + | **[[5iat]] – BaPDD – ''Bacillus anthracis''<br /> | ||
| + | **[[5iax]] – BaPDD + Co+2 + oxoglutarate <br /> | ||
| + | **[[5iav]] – BaPDD + Co+2 + malonate<br /> | ||
| + | |||
| + | * 1,2-dihydroxy-3-keto-methylthiopentene dioxygenase (KMTPD) or acereductone dioxygenase | ||
| + | |||
| + | **[[5i8s]] – mKMTPD + Ni+2 + pentanoate + valerate<br /> | ||
| + | **[[5i8t]] – mKMTPD + Ni+2 + lactate <br /> | ||
| + | **[[5i8y]] – mKMTPD + Co+2 + keto-methylthio-butyrate<br /> | ||
| + | **[[5i91]] – mKMTPD + Ni+2 + keto-methylthio-butyrate<br /> | ||
| + | **[[5i93]] – mKMTPD + Ni+2 + keto-pentanoate<br /> | ||
| + | **[[4qgn]] – hKMTPD + Fe+3<br /> | ||
| + | **[[4qgl]], [[4qgm]] – BaKMTPD + Cd+2<br /> | ||
| + | |||
| + | * Indoleamine 2,3-dioxygenase (IAD) | ||
| + | |||
| + | **[[5ek2]], [[5ek3]], [[5ek4]], [[4pk5]] – hIAD + heme + imidazole derivative<br /> | ||
| + | **[[4u72]], [[4u74]] – hIAD (mutant) + heme + imidazole derivative<br /> | ||
| + | |||
| + | * Thymine dioxygenase (TD) | ||
| + | |||
| + | **[[5c3o]] – NcTD – ''Neurospora crassa''<br /> | ||
| + | **[[5c3p]] – NcTD + oxoglutarate + Ni+2 <br /> | ||
| + | **[[5c3q]] – NcTD + oxoglutarate + Ni+2 + thymine<br /> | ||
| + | **[[5c3s]] – NcTD + oxoglutarate + Ni+2 + pyrimidine derivative <br /> | ||
| + | **[[5c3s]] – NcTD + oxoglutarate + Ni+2 + pyrimidine derivative + uracil derivative<br /> | ||
| + | |||
| + | * 2,4’-dihydroxyacetophenone dioxygenase (APD) | ||
| + | |||
| + | **[[5bpx]], [[4p9g]] – APD + Fe+3 – ''Alcaligenes''<br /> | ||
| + | |||
| + | *Thiol dioxygenase (ThD) | ||
| + | |||
| + | **[[4tlf]] – PaThD + Fe+2 – ''Pseudomonas aeruginosa''<br /> | ||
| + | **[[4wvz]] – PaThD (mutant) + Fe+2 <br /> | ||
}} | }} | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 07:12, 26 April 2017
| |||||||||||
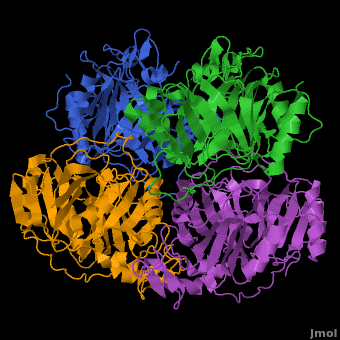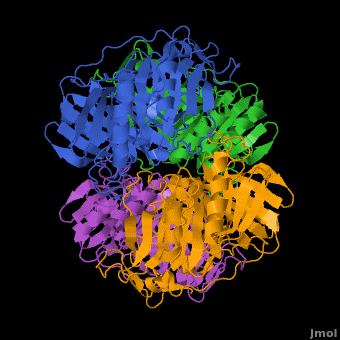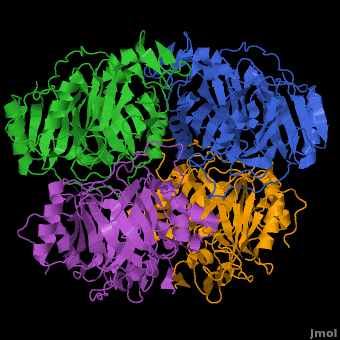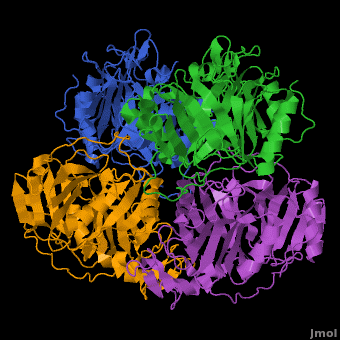
3D structures of Protocatechuate 3,4-dioxygenase
Updated on 26-April-2017
- ↑ Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, Tian Q, Aifantis I, Wei L, Dong C. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015 Apr 21;42(4):613-26. doi: 10.1016/j.immuni.2015.03.005. Epub 2015 , Apr 7. PMID:25862091 doi:http://dx.doi.org/10.1016/j.immuni.2015.03.005
- ↑ Fielding AJ, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L Jr. A hyperactive cobalt-substituted extradiol-cleaving catechol dioxygenase. J Biol Inorg Chem. 2010 Dec 14. PMID:21153851 doi:10.1007/s00775-010-0732-0
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky