Overview
Streptococcus pneumoniae [1] PsaA (Pneumococcal surface antigen A) is a high affinity substrate-binding lipoprotein
[2] detected on all known serotypes of
Streptococcus pneumonia. It facilitates the acquisition of Mn
2+ by being part of the cell's
ABC (ATP binding cassette) transporter and plays a major role in pneumococcal attachment to the host cell and
virulence [3].
Streptococcus pneumoniae, also widely known as pneumococcus, is a Gram positive coccus with a diameter of 0.5 to 1 μm. As a member of the genus Streptococcus it typically forms diplococci and can live both under aerobic or anaerobic conditions. In healthy carriers it resides in the nasopharynx [4]. However, the bacterium may become pathogenic in elderly and immunocompromised people as well as in children due to a weaker immune system. In fact, S. pneumoniae is the most frequent opportunistic pathogen worldwide and causes many types of diseases including otitis media, meningitis, sepsis and pneumonia [5]. The genome of S. pneumoniae is a closed, circular DNA structure that contains between 2.0 and 2.1 million base pairs [4].
PsaA protein
Zinc in excess has a significant toxicity to bacteria because it is an important innate defense mechanism. There are many Zinc molecules in the human body. Manganese is important for the virulence, growth and proliferation of Streptococcus pneumoniae. Zinc could compete for Manganese binding. However Manganese has more affinity for PsaA than Zinc but Zinc is not transported by the ABC-transporter. Zinc competition reduces intracellular Manganese resulting in up-regulation of PsaBCA expression. [6]
Structure of ABC-transporter
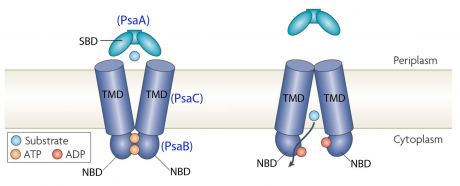
[7] ABC transporter
Bacterial ABC transporters are an important class of transmembrane proteins. They are involved in the import and export of a wide variety of substrates, including sugars, amino acids, peptides, polyamines, and cations [8]. They can be considered as a promising target for antimicrobial strategies. ABC transporters consist of four membrane-associated proteins with two ATP-binding proteins (ATPases) and two permeases. [9]
In the case of PsaA, the ABC transporter is composed of an ATP-binding protein (PsaB), an integral membrane protein (PsaC) and PsaA itself as a lipoprotein that possess a metal-ion binding site. This confers the protein to bind divalent metal-ions, however, with a preference for Mn2+ [10].
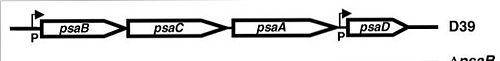
The genes that encode for the components of the ABC transport system are
psaA,
psaB and
psaC, respectively. They are organized consecutively in the psaBCA operon, together with
psaD, the gene encoding for a [thiol peroxidase] (see below).
[11].
General Structure
The protein PsaA has a molecular weight of 34.538 kDa with 309 residues [13]. The overall size of the protein approximated from its crystal structure is 40 by 40 by 70 Å [14].
As a member of the Lipoprotein receptor-associated antigen I (LraI) family, the PsaA molecule contains four distinct regions. An N-terminal leader sequence of 20 amino acids holds an LxACy consensus sequence that is recognized and cleaved by signal peptidase II [14]. A lipid moiety (diacylglycerol [15]) is added to the cysteine residue and mediates the anchorage of the protein to the cytoplasmic membrane. Apart from this leader sequence, the rest of the protein consists of two twofold-pseudosymmetrical (β/α)4 sandwich domains, of which the β-strands of each domain form parallel β-sheets [15]. In total these domains form two lobes connected via an α-helical linker which constitutes the solute-binding site [14].
Secondary structure of PsaA:

Secondary structure of PsaA
[2]
We can see on the right, the 3D structure of PsaA protein. We can observe that there are (TRIS significate 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL).
We can see the . Manganese ions are shown as
.
"3D structure of PsaA with 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL (tris) and cadmium
[16] "
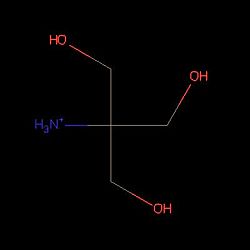
"Structure of Tris PsaA needs to be linked to this molecule to be in an open state"
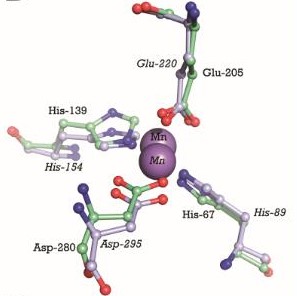
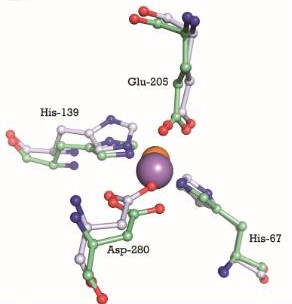
This image shows the metal binding site in more detail, with the MntC residues, manganese (purple) and zinc (orange). We can see that Manganese interacts with Histine residues, Aspartique acid residues and Glutamique acid residues.
The metal binding site is formed by the sidechains of residues His67, His139, Glu205, and Asp280. This site has tetrahedral coordination geometry. The amino acids His67 andHis139 interact with the metal via Nε2 nitrogen atoms. However, the carboxylate sidechains of Glu205 and Asp280 interact with the metal via their Oε1 and Oδ2 atoms. The atomic distance between His67 Nε2 and the metal is 1.99 Ắ while it is 2,01 Ắ for His 139 Nε2. It is 2,04 Ắ for Glu205 Oε1 and 2,02 Ắ for Asp280 Oδ2. The Oδ1 atom of Asp137 can form a hydrogen bond with Nδ1 of His139. Oδ1 atom of Asp65 can also form a hydrogen bond with His67 N. [17]
Cadmium uptake reduces the millimolar cellular accumulation of manganese and zinc, and thereby increases sensitivity to oxidative stress [18].
| 
| 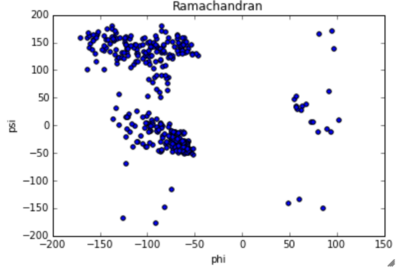 Ramachandran blot of PsaA obtained by Lovell, Davis, et al. Proteins 50:437 |
Function
This protein is essential for the existence of S. pneumoniae as mutations in the psaA gene cause deficiencies in growth but also in its virulence, adherence and the response to oxidative stress.
Adhesin and virulence
PsaA is well hidden beneath the cell wall and the polysaccharide capsule, which is why it does not directly act as an adhesin as initially assumed. This was confirmed by studies on psaB and psaC deletion mutants which were not deficient in PsaA but showed impaired adherence, similar to psaA deletion mutants. As the disruption of either of the members of the Psa operon affects pneumococcal adherence, it seems that this process is coupled to the cellular Mn2+ transport system. With bacterial adhesion being the first step in bacterial pathogenesis, the extinction of PsaA and the associated Psa operon lead to attenuated virulence in affected strains [11].
Virulence
Next to its involvement in bacterial adhesion and the colonization in the human nasopharynx, S. pneumoniae actively produces H2O2 in large amounts to augment its own virulence on the one hand, and, on the other hand, to utilize it as a weapon in the competition against other pathogens [11].
Oxidative stress
In S. pneumoniae H2O2 and superoxides are generated as toxic derivatives of molecular oxygen in several metabolic processes and during growth by pyruvate oxidase. Subsequently, H2O2 reacts with Fe2+ during the Fenton reaction. This results in the formation of the hydroxylyl radical that causes severe damages in the cell. As a protective response against this oxidative stress, S. pneumoniae produces an Mn2+-dependent superoxide dismutase (SodA) and the thiol peroxidase encoded by psaD in the Psa operon. Due to the use of H2O2 during pneumococcal virulence these two proteins and the dependence on Mn2+ are of major importance. When Mn2+ transport is disrupted, bacterial cells display hypersensitivity towards oxidative stress [11].
Mn2+ enables Streptococcus Pneunomiae to have a prompt and rapid growth. Superoxides are a key player in oxidative stress generated in the presence of iron. Therefore, they inhibit growth in the absence of Mn2+.
Mn2+ is also required for the activity of CpsB, a tyrosine phosphatase involved in the regulation of capsule production. And in some streptococcal species, lectin-mediated adherence requires Mn2+. Finally, mutations in the psa operon result in an almost complete attenuation of virulence for all tested models of animal infection.
Interactions with other proteins
- piaA, pneumococcal iron acquisition A which is an iron-compound ABC transporter permease. It is required for bacterial growth and full virulence in pulmonary infection.
-pavA, Pneumococcal adherence and virulence factor A is an adherence and virulence protein A. It binds fibronectin.
-Wzg, which regulates the capsule production and is an integral membrane regulatory protein Cps2A
-Tpx, a thiol peroxidase with a Has antioxidant activity. This protein can remove peroxides or H2O2 by similarity.
-PsaC, a manganese ABC transporter permease
-PsaB, a manganese ABC transporter ATP-binding protein. Deleting psaB results in decreased expression of PsaA. PsaC and psaC require Mn2+ supplementation for growth and transformation.
-SPD_1450, an iron-dependent transcriptional regulator
-adcC, antibody-dependent cell-mediated cytotoxicity is a zinc ABC transporter ATP-binding protein.
-adcB, a zinc ABC transporter permease
Disease
Otitis media
PsaA has been recognized to be involved in the adherence and virulence mechanisms of otitis media. Streptococcus pneumoniae is one of the main agents causing bacterial acute otitis media, directly or as complication of a viral upper respiratory tract infection. This disease is a highly prevalent pediatric disease worldwide. Hearing loss is a common problem associated with this disease. Streptococcus pneumoniae in middle ear can be resistant to penicillin and cephalosporin[19]
Pneumonia
PsaA protein is an indirect adhesin which is involved in colonization of the nasopharyngeal mucosal. Moreover, alveolar pneumonia is caused by the spread of Streptococcus pneumoniae from nasopharynx. Therefore PsaA protein is involved in infection of pulmonary parenchyma by Streptococcus pneumoniae.
Sometimes, Streptococcus pneumoniae pass into the blood and causes a bacteremia besides pneumonia.
Application in Biotechnology
PsaA is being actively evaluated as a component of a vaccin in formulations composed of pneumococcal common proteins. PsaA is a component of a vaccin because this protein is immunogenic and stimulates an increase in antibody production when the nasopharynx is naturally colonized. PsaA has been expressed as an E.coli recombinant protein, purified, and evaluated in a phase one clinical trial.
Progress in vaccine development is most advanced for Streptococcus pneumoniae. Indeed, there is a seven-valent capsular-conjugate vaccine, PREVNAR® but it is rather non efficient for otitis media.[20] PsaA has been shown to be an effective vaccine in animal models at preventing Streptococcus pneumoniae infection.
References
- ↑ http://www.sciencephoto.com/images/download_lo_res.html?id=662360183
- ↑ 2.0 2.1 http://www.uniprot.org/uniprot/P0A4G2
- ↑ Bajaj M, Mamidyala SK, Zuegg J, Begg SL, Ween MP, Luo Z, Huang JX, McEwan AG, Kobe B, Paton JC, McDevitt CA, Cooper MA. Discovery of novel pneumococcal surface antigen A (PsaA) inhibitors using a fragment-based drug design approach. ACS Chem Biol. 2015 Jun 19;10(6):1511-20. doi: 10.1021/cb501032x. Epub 2015 Mar, 30. PMID:25786639 doi:http://dx.doi.org/10.1021/cb501032x
- ↑ 4.0 4.1 Bierman EL, Stein O, Stein Y. Lipoprotein uptake and metabolism by rat aortic smooth muscle cells in tissue culture. Circ Res. 1974 Jul;35(1):136-50. PMID:4366526
- ↑ Deplazes E, Begg SL, van Wonderen JH, Campbell R, Kobe B, Paton JC, MacMillan F, McDevitt CA, O'Mara ML. Characterizing the conformational dynamics of metal-free PsaA using molecular dynamics simulations and electron paramagnetic resonance spectroscopy. Biophys Chem. 2015 Dec;207:51-60. doi: 10.1016/j.bpc.2015.08.004. Epub 2015 Sep, 3. PMID:26379256 doi:http://dx.doi.org/10.1016/j.bpc.2015.08.004
- ↑ McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog. 2011 Nov;7(11):e1002357. Epub 2011 Nov 3. PMID:22072971 doi:10.1371/journal.ppat.1002357
- ↑ http://www.latrobe.edu.au/biochemistry-and-genetics/research/maher/psabca-manganese-uptake-by-streptococcus-pneumoniae
- ↑ Tagawa M, Sakamoto T, Tamura Y, Imai K, Ito T, Matsubara H, Kanno M, Shigemoto K, Koseki H, Taniguchi M. Genomic DNA with transformation-related activity and melanoma antigen expression. J Invest Dermatol. 1989 May;92(5 Suppl):284S-288s. PMID:2715661
- ↑ http://webcache.googleusercontent.com/search?q=cache:3MvkKDHQVzwJ:www.omicsonline.com/open-access/role-of-abc-transporter-proteins-in-stress-responses-of-streptococcus-mutans-2247-2452.1000592.pdf+&cd=1&hl=fr&ct=clnk&gl=fr&client=firefox-b
- ↑ Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008 Apr;6(4):288-301. doi: 10.1038/nrmicro1871. PMID:18340341 doi:http://dx.doi.org/10.1038/nrmicro1871
- ↑ 11.0 11.1 11.2 11.3 Moore RC. Investigational drug information is available to the pharmacist. Am J Hosp Pharm. 1979 Nov;36(11):1480, 1484. PMID:517531
- ↑ McAllister LJ, Tseng HJ, Ogunniyi AD, Jennings MP, McEwan AG, Paton JC. Molecular analysis of the psa permease complex of Streptococcus pneumoniae. Mol Microbiol. 2004 Aug;53(3):889-901. PMID:15255900 doi:http://dx.doi.org/10.1111/j.1365-2958.2004.04164.x
- ↑ https://www.mybiosource.com/prods/Recombinant-Protein/Manganese-ABC-transporter-substrate-binding-lipoprotein-psaA/psaA
- ↑ 14.0 14.1 14.2 Yu Y, Chang, Xu H, Zhang X, Pan L, Xu C, Huang B, Zhou H, Li J, Guo J, Liu C. The virulence of Streptococcus pneumoniae partially depends on dprA. Braz J Microbiol. 2016 Dec 6. pii: S1517-8382(16)30151-4. doi:, 10.1016/j.bjm.2016.10.019. PMID:28011228 doi:http://dx.doi.org/10.1016/j.bjm.2016.10.019
- ↑ 15.0 15.1 Albrecht P. Immune control in experimental subacute sclerosing panencephalitis. Am J Clin Pathol. 1978 Jul;70(1 Suppl):175-84. PMID:99024
- ↑ http://www.ebi.ac.uk/pdbe/entry/pdb/4UTO
- ↑ Lawrence MC, Pilling PA, Epa VC, Berry AM, Ogunniyi AD, Paton JC. The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure. 1998 Dec 15;6(12):1553-61. PMID:9862808
- ↑ http://www.nature.com/articles/ncomms7418
- ↑ https://www.google.com/patents/US20130078254
- ↑ Wald ER, Mason EO Jr, Bradley JS, Barson WJ, Kaplan SL. Acute otitis media caused by Streptococcus pneumoniae in children's hospitals between 1994 and 1997. Pediatr Infect Dis J. 2001 Jan;20(1):34-9. PMID:11176564